
The association between cytochrome P450 polymorphisms and anti-tuberculosis drug-induced liver injury: a systematic review and meta-analysis
Introduction
Tuberculosis (TB) is a worldwide infectious disease caused by Mycobacterium tuberculosis, which takes more lives than all other infectious diseases. Drug-resistant TB poses a serious threat to public health systems all over the world. Owing to the forward steps made in treating and preventing TB, in 2019, it is estimated that there will be 9.96 million new TB cases and 1.41 million deaths in the world. Since 2007, TB has been ranked as one of the top ten causes of death in the world and ranked first among all infectious diseases (1). Although novel diagnostic methods, medications, and vaccination for TB have been trialed, the most recommended drugs for the first-line treatment of TB are still isoniazid (INH), rifampicin (RMP), pyrazinamide (PZA), and ethambutol (EMB) with/without streptomycin. Chemotherapy drugs which can effectively treat TB are widely available, but their side effects cannot be overlooked, with the severest being anti-tuberculosis drug-induced liver injury (ADTILI). If the patient has ADTILI but fails to identify and stop treatment in time, the patient may have serious adverse reactions or even death.
N-acetyltransferase 2 (NAT2) initially metabolizes INH into acetylisoniazid, which is subsequently hydrolyzed to acetylhydrazine. NAT2 was further acetylated to produce a non-toxic metabolite diacetylhydrazine. Acetylhydrazine can also be oxidized by cytochrome P450 2E1 (CYP2E1), generating hepatotoxins including acetyldiazene, acetylonium ion, acetyl radical, and ketene. More than NAT2 and CYP2E1, a relationship may potentially exist between polymorphisms in genes coding for the microsomal cytochrome P450 (CYP) enzymes, such as CYP3A4, and ATDILI. RMP induces CYP, resulting the level of toxic products increased after INH metabolism (2). The pregnane X receptor (PXR) closely modulates gene expression in the hepatic drug-clearance system, and its unwanted activation can contribute to ATDILI (3). RMP mediates the activation of PXR, which operates as a pivotal factor which controls xenobiotic and drug-mediated induction of the CYP3A, CYP2B, and CYP2C subfamilies (4). CYP enzymes may be of crucial importance to reactive metabolite synthesis and detoxification (5). Researchers have concentrated efforts toward depicting the association between ATDILI and CYP enzymes, such as CYP2B6 (6), CYP2C9, CYP2C19 (7), and CYP2D6 (8); however, their results have been controversial and contradictory.
To our knowledge, four meta-analyses regarding ATDILI’s relationship with CYP2E1 RsaI/PstI polymorphisms have been conducted previously (9-12). However, these studies have some inadequacies; for instance, none of them included studies published after 1st August, 2015, and none of them reviewed any other CYPs. In the present work, we systematically reviewed and meta-analyzed all published articles examining the extent of the supposed genetic relationship between CYP polymorphism and susceptibility to ATDILI.
We present the following article in accordance with the PRISMA reporting checklist (available at https://dx.doi.org/10.21037/apm-21-1224).
Methods
Search strategy
We performed searches of Medline, Embase, the Cochrane Library, Web of Science, and Chinese National Knowledge Infrastructure (CNKI) to identify relevant studies written in Chinese or English and published before June 11, 2018. The following Medical Subject Headings (MeSH) terms were used as keywords for our searches: “cytochrome P450 enzyme”, “drug-induced liver injury”, “polymorphism”, "tuberculosis”, and “hepatotoxicity”. We also manually screened the references from published meta-analyses to identify any other literature of potential interest.
Eligibility screening
To be included in the meta-analysis, studies needed to meet the following criteria: (I) the study was published in peer-reviewed journals and the original data were available; (II) the study included details of genotypic distribution of polymorphisms for the ATDILI groups and the control group; (III) the type of study design as a case-control or cohort study; and (IV) the details of the included studies included the definition of ATDILI and the detailed treatment plan of the patients. The exclusion criteria were: (I) published literature repeatedly; (II) reviews and meta-analyses; (III) animal research; (IV) no association with CYPs.
Data extraction and assessment of study quality
Two authors extracted information from each article independently, including: the name of the first author; publication year; study participants’ age, sex, and ethnicity; diagnostic criteria; genotyping method; numbers of participants in the case group and control group; and the frequency of genotypes among the ATDILI and control groups. We contacted the authors directly if the study participants’ chemotherapy protocol were not mentioned in the studies. A response was received from one query, and the article was included (13). If there were differences on the included articles, the two authors discussed and decide whether to include. We assessed the quality of the studies based on the criteria revised by Deng (11).
Statistical analyses
CYP2E1 genotypes were analyzed according to the genetic model of proposed risk (C1/C1 vs. C1/C2 and C2/C2 for the RsaI/PstI polymorphism). All data were statistically analyzed with STATA version 14.0 (Stata, College Station, TX, USA) and Review Manager, version 5.3 (Revman, Cochrane Centre, The Cochrane Collaboration). The standard Q-statistic test was adopted to assess the studies for heterogeneity. Heterogeneity was evidenced by I2>50%. If heterogeneity was present, we would employ the fixed-effects model; in its absence, the random-effects model would be adopted. Summary odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to analyze the correlation of genotypes with the risk of ATDILI in patients, with P<0.05 being an indication of statistical significance. We conducted subgroup analyses according to: (I) ethnicity (East Asian, South Asian, or South American); (II) chemotherapy protocol; (III) the study design: cohort or case-control; and (IV) the definition of ATDILI based on a minimum serum alanine aminotransferase (ALT) level the upper limit of normal (ULN). We tested the study results for stability by performing a sensitivity analysis, and Begg’s funnel plot was used for publication bias detection.
Results
Study characteristics
From the 116 articles originally identified in our searches, 38 eligible studies were finally included. After reading these papers carefully, we found that 2 articles had duplicated data. Therefore, 37 studies described the relationship between CYP polymorphism and susceptibility to ATDILI, of which 30 studies were related to CYP2E1 (2 focused on both CYP2E1 and other CYPs), and 7 studies focused on the distribution of other CYPs. The selection process for the eligible studies is depicted in Figure 1. Among the 30 articles focusing on CYP2E1, 29 articles discussed CYP2E1 RsaI/PstI polymorphisms and ATDILI, and 1 article discussed other tag single-nucleotide polymorphisms (SNPs) in CYP2E1 (14). Further, 4 articles included participants with CYP3A4 (6,7,15,16), 2 articles included participants with, CYP2B6 (6,17), 2 articles with CYP2C9 (7,18), 2 articles with CYP2C19 (7,15,19), and 3 articles with CYPC3A5 (6,15,18) polymorphisms, respectively.
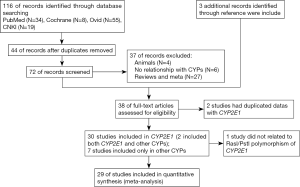
Eventually, 29 studies involving 7,526 patients (1,548 in the case group and 5,978 in the control group) satisfied the criteria for inclusion and were subsequently meta-analyzed. Among them, 11 articles included participants of Chinese ethnicity (8,14,20-29), 7 articles included Indian participants (30-35), 5 articles included Brazilian participants (2,13,36-38), 2 included Korean participants (39,40), 3 articles included participants of more than 1 ethnicity (41-43), 1 included Indonesian participants (44), 1 included Japanese participants (45), and 1 included Tunisian participants (46). Polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) was the method used most frequently to detect genotypes. Nine articles were case-control studies, and 19 were cohort studies (of which 1 was retrospective, and the others were prospective); the design method is not clear in one study
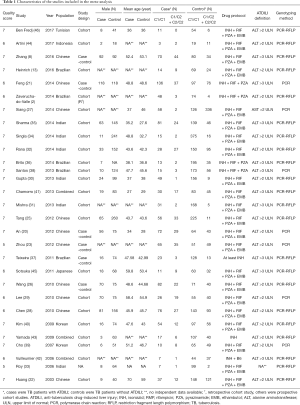
The main characteristics were summarized in Table 1. Of the patients who underwent the anti-TB treatment 1,548 case were with ADTIH, while 5,978 were not.
Full table
Association of CYP2E1 gene with ATDILI
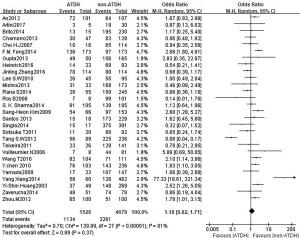
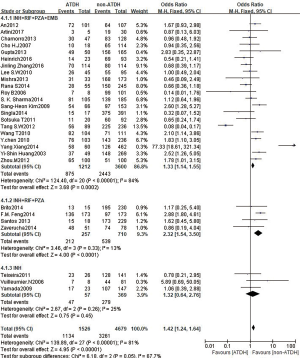
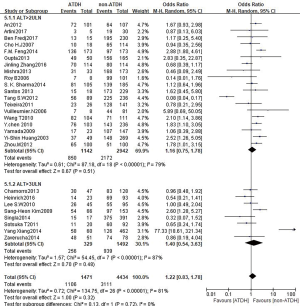
A total of 28 studies reported an association between the CYP2E1 gene and ATDILI, and there was obvious heterogeneity among the studies (χ2=139.89, I2=81%, P<0.05); consequently, we adopted the random-effects approach for the meta-analysis. The pooled OR of all studies for CYP2E1 to the risk of ATDILI was 1.18 (95% CI: 0.82–1.71, P=0.37, Figure 2). This result showed that CYP2E1 RsaI/PstI polymorphisms were not statistically associated with ATDILI. We further performed subgroup analysis according to ethnicity, with the ethnic groups in the studies mainly including East Asians, South Asians, and South Americans. However, There was no significant difference between subgroups (P=0.31, Figure 3). Subgroup analysis was performed according to the participants’ drug protocol. The results showed that the patients who receiving a four-drug protocol (INH + RIF + PZA + EMB) or three-drug protocol (INH + RIF + PZA) regimens showed a higher risk of ATDILI than those who receiving INH alone (OR =1.33, 95% CI: 1.14–1.55; OR =2.32, 95% CI: 1.54–3.50) and OR =1.32, 95% CI: 0.64–2.76, respectively) (Figure 4). Additionally, we analyzed the subgroups according to study type (cohort design or case-control) and the definition of ATDILI (ALT >3 ULN or ALT >2 ULN); however, no significant difference was found between the subgroups (Figures 5,6).


Sensitivity analysis and publication bias
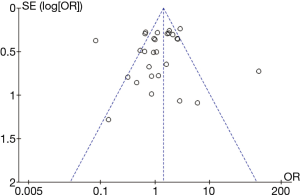
Low-quality studies were excluded in sensitivity analysis to determine whether the literature quality was the source of heterogeneity. Sensitivity analysis after the elimination of low quality literature (score <6) was carry out. We found that there was still extremely obvious heterogeneity between the studies, indicating that the source of heterogeneity was not the quality of the literature. Begg’s funnel plot analysis of the 29 included studies concerning CYP1E2 and ATDILI suggested an acceptable level of publication bias (t=1.60, P=0.122) (Figure 7).
Associations of other genes in the CYP family with ATDILI
Nine articles focused on the relationships of other genes in the CYP family with ATDILI. Although the heterogeneity among the articles was significant, there was some consistency. For instance, the 516 TT homozygous mutant of CYP2B6 was used for the risk analysis. One study base on Chinese population suggested male patients harboring the CYP2B6 *6/*6 genotype to potentially have lower susceptibility to developing ATDILI than female (P=0.039, OR =0.097, 95% CI: 0.011–0.885) (17). In Another study found the 516 TT homozygous mutant genotype of the CYP2B6 gene to also carry significance (P=0.046; OR =0.063, 95% CI: 0.004–0.955) in a key variable analysis (6).
With respect to CYP3A4, the situation was even more indistinct. For the CYP3A4 gene (c.-392 G>A) CYP3A4 genotypes were not significantly associated with ADTILI in Zaverucha-do-Valle’s study (2), which was most likely attributable to most of the participants in the analysis carrying the homozygous wild-type genotype. Meanwhile, in Sun’s study, the CYP3A4 18B20332G/A genotype was found to be a protective genotype against ADTILI (OR =0.088 95% CI: 0.027–0.291) (15). Moreover, in Guo’s study, the CYP3A4 18B genotype elevated the risk of developing ADTILI (OR =1.196, 95% CI: 1.231–2.980) (16).The results were equivocal. The main characteristics and genotypes are summarized in Tables 2,3, respectively.
Full table

Full table
Discussion
In the present study, we meta-analyzed 29 studies involving 7,526 cases (1,548 with ATDILI, and 5,978 without ATDILI) which explored the association of CYP2E1 RsaI/PstI gene polymorphism with ADTILI. Our study found that RsaI/PstI polymorphisms in the CYP2E1 gene were associated with an increased risk of developing ATDILI among East Asian populations. This conclusion was in agreement with the results of three other meta-analyses (47).INH showed a reduced inhibitory effect on CYP2E1 activity among patients carrying the CYP2E1 RsaI/PstI c1/c1 genotype compared to patients with other genotypes. Thus, as a result of INH treatment, participants harboring the CYP2E1 c1/c1 genotype display increased CYP2E1 activity compared to patients carrying other genotypes; consequently, these patients may display elevated hepatotoxin production, increasing their chances of sustaining liver injury (10).
The human derived PXR, also known as nuclear receptor 1l2, was initially considered as the main regulatory factor of exogenous detoxification. It regulates the expression of drug metabolizing enzymes and transporters to control the degradation and excretion of exogenous and endogenous substances (including therapeutic drugs). PXR gene polymorphism is considered to be related to adverse reactions and interactions of drugs (or exogenous drugs) (48). The polymorphism of PXR gene and anti-tuberculosis drug-induced hepatotoxicity (ATDH) susceptibility have attracted the attention of researchers. In 2015, Zazuli et al. analyzed the PXR gene polymorphism of 106 Indonesian people, and found that the T genotype of rs3814055 was related to high risk of ATDH, so it was considered that PXR gene polymorphism was one of the risk factors of ATDH (49). In the same year, Wang et al. analyzed the polymorphism of PXR combined NAT gene in 355 Taiwan people at the same time. It was found that NAT2 and malnutrition were independent risk factors of ATDH in male and female patients, but AA genotype of rs2461823 and an allele of rs6785049 were only independent risk factors for female sex. The results show that SNP genotype and haplotype of PXR are still important risk factors of ATDH in Asian population, and show the characteristics of gender stratification (50). In 2019, Wang et al. studied 502 Chinese TB patients and found that the secondary allele and h0010001 haplotype of rs7643645 in PXR were related to reducing the risk of ATDH, and suggested that the drug metabolic enzyme regulated by PXR was related to the pathogenesis of ATDH (51). In 2019, Zhang et al. conducted allele frequency, genotype and genetic model of PXR gene in 746 patients with TB in Western China, and analyzed interaction with SNP. It was found that the risk of ATDH in T allele carriers of rs3814055 was lower than that of C allele carriers, the dominant and additive models in the main genetic models also suggested that the locus was related to the risk of ATDH susceptibility (52).
NAT2 and CYP2E1 are the key enzymes for the detoxification of INH in liver (5). Due to the association between the “slow acetylation” phenotype of NAT2 gene and the increased blood level and toxic reaction rate of INH, FDA has included the NAT2 gene phenotype in the drug label of INH for the treatment of TB (53). PharmGKB database also uses important NAT2 genetic variation as a clinical guideline to predict the risk of ATDH induced by INH in TB patients. Researchers have studied a large number of gene polymorphisms in drug metabolism, such as CYP2E1, glutathione S-transferase (GST), cholesterol 7α-hydroxylase gene, bile salt export pump (BSEP) gene, sodium ion sodium taurocholate cotransporter polypeptide gene and PXR gene. hydrazine (Hz), acetyldiazine (AC) and their metabolites, which are the toxic metabolites of INH, can cause liver injury, which is related to the metabolism of NAT2, CYP2E1 and GST had correlation; RIF also has weak hepatotoxicity due to the enhancement of INH hydrolase activity and/or activation of PXR, such as CYP, glutathione, bile acid and lipid metabolism enzymes, with the incidence of less than 1.1% (3,54,55).
The occurrence and development of ATDH involves many complex links, such as drug metabolism, oxidative stress, mitochondrial dysfunction, immune regulation and inflammatory response. These links occur simultaneously or sequentially, and the interaction of each link determines the occurrence, outcome and prognosis of ATDH. Drug metabolic transport is the initiation of ATDH, especially the metabolism of INH is related to the metabolic type of metabolic enzyme NAT (5,53,56).
Drug metabolizing enzyme gene polymorphism has always been the research direction of ATDH, but most of them are small sample and single ethnic analysis, and there is no recognized conclusion about the correlation between NAT gene polymorphism and ATDH (53).
Studies in the case-control design subgroup seemed to show high risks of ATDILI. To some degree, this result may be explained by the accurate matching in advance, which eliminated some mixed factors, such as age and sex.
Both epigenetic and genetic alterations may affect the expression of GSTP1 and CYP1A1, such as abnormal changes in CpG island methylation of the GSTP1 promoter. Gene promoter methylation is generally related to transcriptional repression via mechanisms such as direct prevention of transcription factor binding to DNA binding sites or via complex indirect mechanisms such as chromatin remodeling. Toxic metabolites of anti-TB drugs may induce the methylation of the promoter regions of GSTP1 or CYP1A1 (19).
Down-regulated expression of bile acid transporters, BSEP, and sodium taurocholate co-transporting polypeptide (NTCP) are found in mice co-treated with INH and RMP, which is associated with an increased risk of the ATDILI (28). Increased expression of CYP7A1 occurs in Wistar rats co-treated with INH and RMP, which results in excess bile acids based on histo-pathological studies (29).Therefore, alterations of CYP7A1 and BSEP in the accumulation of bile acids may contribute to ATDILI (53).
We also found that the risk of ATDILI varies with different treatment protocol for TB. Only three studies used INH as the sole anti-TB treatment, while most of the others used a three- or four-drug combination regimen. Thus, it was equivocal that INH alone seemed to have a lower risk (OR =0.97), while combination regimens seemed to have a higher risk. In most of the articles, the definition of ATDILI was an ALT level of at least two- or three-fold the ULN, with or without an elevated level of aspartate aminotransferase (AST), and with or without symptoms of hepatitis. We stratified ATDILI into subgroups only by levels of ALT, and found a statistical association between ATIDIL risk and the ALT >2 ULN subgroup (OR =1.35, 95% CI: 1.05–1.74, P=0.021); however, taking the heterogeneity into consideration (I2=44.4%, P=0.02), the analysis maybe under power. It is worth mentioning that heterogeneity dramatically decreased from I2=44% to I2=23%, following the removal of Feng et al.’s article (21), with a seemingly stable association (OR =1.26, 95% CI: 1.01–1.58, P=0.04).
In our study, a total of 28 studies reported an association between the CYP2E1 gene and ATDILI, and there was obvious heterogeneity among the studies (χ2=139.89, I2=81%, P<0.05); The pooled OR of all studies for CYP2E1 to the risk of ATDILI was 1.18 (95% CI: 0.82–1.71, P=0.37, Figure 2). This result showed that CYP2E1 RsaI/PstI polymorphisms were not statistically associated with ATDILI. Our meta-analysis has uncovered an association between CYP2E1 RsaI/PstI polymorphisms and ATDILI, especially among patients who receive a four-drug (INH + RIF + PZA + EMB) or three-drug (INH + RIF + PZA) anti-TB treatment regimen.
The Begg’s funnel plot (P=0.122) showed the included studies to have an acceptable level of publication bias. Compared with previous meta-analyses, there was a remarkable increase in articles with a larger number of cases with different origins and over a long-time span. It is possible that studies based on smaller sample sizes could have tilted toward producing positive results. Second, as in Feng et al.’s study (21), liver function detection was done at 6 months after the end of therapy, which is a later point in time than in most of the reported studies (6,17,21). A large proportion of studies reported that ADTILI occurred within 2 months after the end of treatment, which suggests there was potential selection bias. Third, there were many case-control studies in this research, which may have contributed to selection bias. Finally, we only do meta-analysis on those published articles. Therefore, mild publication bias potentially existed.
There are also some limitations to our meta-analysis. First, we did not research DraI polymorphisms in the CYP2E1 gene, as they have rarely been reported on since the publication of Wang’s study (12); moreover, none of the existing meta-analyses declared any association between DraI polymorphism and ADTILI. Second, we did not evaluate the potential association between NATs and CYP2E1, and there may be combined effects of these two genes. Further analyses should be conducted to assess the combination of NAT statues and CYP2E1 genotypes on the risk of ATDIL. Third, there are many other high-risk factors for ATIDLI, such as alcohol consumption, sex, diabetes, malnutrition, HIV infection and viral hepatitis (5).Although it is difficult to explore these underlying risk factors in a simple meta-analysis, the gene-to-gene, gene-to-environment, and gene phenotypes should not be neglected. Finally, there was a lack of evidence of other CYPs related to ATIDIL due to the limited number of studies, and even in the handful of studies available, the authors did not focus on the same tag SNPs.
In the transcription factor binding sites of the PXR regulatory region (the promoter and intron 1) have also been associated with PXR alteration and CYP3A4 expression, and drug-induced liver injury (50). As RIF is a human PXR–specific activator, it is reasonable to believe that the polymorphism of CYP3A4 may have an effect on RIF-induced liver injury, although to date, there have been no positive findings (7).
Notably, in Zhang’s research (8), a correlation was identified between CpG island hypermethylation of the CYP2E1 and CYP2D6 genes and the development of ADTILI. This finding may bring new insight into the relationship between the epigenetic candidate genes with ADTILI.
In conclusion, the results of this meta-analysis indicate an association between CYP2E1 genetic polymorphisms and ATDILI, especially in patients treated with a four-drug (INH + RIF + PZA + EMB) or three-drug (INH + RIF + PZA) anti-TB treatment regimen. Genetic mutations of CYP enzyme family members and alterations in DNA methylation levels are perhaps only two of many risk factors for ATDILI, with other extrinsic factors waiting to be discovered, including diet, alcohol consumption, smoking, existing liver disease, and other co-existing diseases. Furthermore, the gene-to-gene, gene-to-environment, and host immunity against varied strains of TB also need to be explored.
Acknowledgments
Funding: This project was supported by the National Natural Science Foundation of China (grant no. 81700581), the Project of Science and Technology Department of Sichuan Province (grant no. 2019YFH0069), Chengdu Medical Research Project (grant no. 2020208), and Sichuan Provincial People’s Hospital Clinical Research and Transformation Fund (2018LY12).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-1224
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-1224). All authors report that this project was supported by the Project of Science and Technology Department of Sichuan Province (grant no. 2019YFH0069), Chengdu Medical Research Project (grant no. 2020208), and the Project of Sichuan Medicine Research (S20014). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chakaya J, Khan M, Ntoumi F, et al. Global Tuberculosis Report 2020 - Reflections on the Global TB burden, treatment and prevention efforts. Int J Infect Dis 2021; Epub ahead of print. [Crossref] [PubMed]
- Zaverucha-do-Valle C, Monteiro SP, El-Jaick KB, et al. The role of cigarette smoking and liver enzymes polymorphisms in anti-tuberculosis drug-induced hepatotoxicity in Brazilian patients. Tuberculosis (Edinb) 2014;94:299-305. [Crossref] [PubMed]
- Wang YM, Chai SC, Brewer CT, et al. Pregnane X receptor and drug-induced liver injury. Expert Opin Drug Metab Toxicol 2014;10:1521-32. [Crossref] [PubMed]
- Chang HY, Chen CJ, Ma WC, et al. Modulation of pregnane X receptor (PXR) and constitutive androstane receptor (CAR) activation by ursolic acid (UA) attenuates rifampin-isoniazid cytotoxicity. Phytomedicine 2017;36:37-49. [Crossref] [PubMed]
- Ramappa V, Aithal GP. Hepatotoxicity related to anti-tuberculosis drugs: mechanisms and management. J Clin Exp Hepatol 2013;3:37-49. [Crossref] [PubMed]
- Fernandes DC, Santos NP, Moraes MR, et al. Association of the CYP2B6 gene with anti-tuberculosis drug-induced hepatotoxicity in a Brazilian Amazon population. Int J Infect Dis 2015;33:28-31. [Crossref] [PubMed]
- Tang SW, Lv XZ, Chen R, et al. Lack of association between genetic polymorphisms of CYP3A4, CYP2C9 and CYP2C19 and antituberculosis drug-induced liver injury in a community-based Chinese population. Clin Exp Pharmacol Physiol 2013;40:326-32. [Crossref] [PubMed]
- Zhang J, Zhu X, Li Y, et al. Correlation of CpG island methylation of the cytochrome P450 2E1/2D6 genes with liver injury induced by anti-tuberculosis drugs: a nested case-control study. Int J Environ Res Public Health 2016;13:776. [Crossref] [PubMed]
- Sun F, Chen Y, Xiang Y, et al. Drug-metabolising enzyme polymorphisms and predisposition to anti-tuberculosis drug-induced liver injury: a meta-analysis. Int J Tuberc Lung Dis 2008;12:994-1002. [PubMed]
- Cai Y, Yi J, Zhou C, et al. Pharmacogenetic study of drug-metabolising enzyme polymorphisms on the risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. PLoS One 2012;7:e47769 [Crossref] [PubMed]
- Deng R, Yang T, Wang Y, et al. CYP2E1 RsaI/PstI polymorphism and risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. Int J Tuberc Lung Dis 2012;16:1574-81. [Crossref] [PubMed]
- Wang FJ, Wang Y, Niu T, et al. Update meta-analysis of the CYP2E1 RsaI/PstI and DraI polymorphisms and risk of antituberculosis drug-induced hepatotoxicity: evidence from 26 studies. J Clin Pharm Ther 2016;41:334-40. [Crossref] [PubMed]
- Heinrich MM, Zembrzuski VM, Ota MM, et al. Factors associated with anti-TB drug-induced hepatotoxicity and genetic polymorphisms in indigenous and non-indigenous populations in Brazil. Tuberculosis (Edinb) 2016;101:15-24. [Crossref] [PubMed]
- Sun Q, Liu HP, Zheng RJ, et al. Genetic polymorphisms of SLCO1B1, CYP2E1 and UGT1A1 and susceptibility to anti-tuberculosis drug-induced hepatotoxicity: a Chinese population-based prospective case-control study. Clin Drug Investig 2017;37:1125-36. [Crossref] [PubMed]
- Sun SF, Li B, Chong YZ, et al. Relationships of CYP450, GSTs, and UGT gene polymorphisms with anti-tuberculosis drug-induced liver injury. Shandong Medical Journal 2017;57:6-10.
- Guo TJ, Li YH, Zhu LY, et al. Association between CYP3A5*3/CYP3A4*18-B gene polymorphisms and liver injury induced by anti-tuberculosis drugs. Chinese Journal of Disease Control & Prevention 2016;20:897-900, 909.
- Wang Y, Xiang X, Wu SQ, et al. Association of CYP2B6 gene polymorphisms and anti-tuberculosis drug-induced hepatotoxicity in a Chinese population. Infect Genet Evol 2017;51:198-202. [Crossref] [PubMed]
- He L, Gao L, Shi Z, et al. Relationship study between the gene polymorphisms of cytochrome P450 1A2/2C19 and anti-tuberculosis drug-induced hepatic injury. Chinese Pharmaceutical Journal 2015;50:248-52.
- He L, Gao L, Shi Z, et al. Involvement of cytochrome P450 1A1 and glutathione S-transferase P1 polymorphisms and promoter hypermethylation in the progression of anti-tuberculosis drug-induced liver injury: a case-control study. PLoS One 2015;10:e0119481 [Crossref] [PubMed]
- An HR, Wu XQ, Wang ZY, et al. NAT2 and CYP2E1 polymorphisms associated with antituberculosis drug-induced hepatotoxicity in Chinese patients. Clin Exp Pharmacol Physiol 2012;39:535-43. [Crossref] [PubMed]
- Feng FM, Guo M, Chen Y, et al. Genetic polymorphisms in metabolic enzymes and susceptibility to anti-tuberculosis drug-induced hepatic injury. Genet Mol Res 2014;13:9463-71. [Crossref] [PubMed]
- Huang YS, Chern HD, Su WJ, et al. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology 2003;37:924-30. [Crossref] [PubMed]
- Zhou M, Li FJ, Zhang L, et al. Study on Chinese medicine constitution and its relation with CYP2E1 gene polymorphisms of anti-tuberculosis drug-induced hepatitis. Chinese Archives of Traditional Chinese Medicine 2012;30:1841-3.
- Tang S, Lv X, Zhang Y, et al. Cytochrome P450 2E1 gene polymorphisms/haplotypes and anti-tuberculosis drug-induced hepatitis in a Chinese cohort. PLoS One 2013;8:e57526 [Crossref] [PubMed]
- Tang SW, Lv XZ, Zhang Y, et al. CYP2E1, GSTM1 and GSTT1 genetic polymorphisms and susceptibility to antituberculosis drug-induced hepatotoxicity: a nested case-control study. J Clin Pharm Ther 2012;37:588-93. [Crossref] [PubMed]
- Wang T, Yu HT, Wang W, et al. Genetic polymorphisms of cytochrome P450 and glutathione S-transferase associated with antituberculosis drug-induced hepatotoxicity in Chinese tuberculosis patients. J Int Med Res 2010;38:977-86. [Crossref] [PubMed]
- Xiang Y, Ma L, Wu W, et al. The incidence of liver injury in Uyghur patients treated for TB in Xinjiang Uyghur autonomous region, China, and its association with hepatic enzyme polymorphisms nat2, cyp2e1, gstm1 and gstt1. PLoS One 2014;9:e85905 [Crossref] [PubMed]
- Chen Y, Guo M, Li SM, et al. Study of the relationship between polymorphisms of cytochrome P450 2E1 and antituberculosis drug-induced hepatic injury. Chinese Journal of Infectious Diseases 2010;28:748-52.
- Lee SW, Chung LS, Huang HH, et al. NAT2 and CYP2E1 polymorphisms and susceptibility to first-line anti-tuberculosis drug-induced hepatitis. Int J Tuberc Lung Dis 2010;14:622-6. [PubMed]
- Gupta VH, Amarapurkar DN, Singh M, et al. Association of N-acetyltransferase 2 and cytochrome P450 2E1 gene polymorphisms with antituberculosis drug-induced hepatotoxicity in Western India. J Gastroenterol Hepatol 2013;28:1368-74. [Crossref] [PubMed]
- Mishra S, Daschakraborty S, Shukla P, et al. N-acetyltransferase and cytochrome P450 2E1 gene polymorphisms and susceptibility to antituberculosis drug hepatotoxicity in an Indian population. Natl Med J India 2013;26:260-5. [PubMed]
- Rana SV, Sharma SK, Ola RP, et al. N-acetyltransferase 2, cytochrome P4502E1 and glutathione S-transferase genotypes in antitubercular treatment-induced hepatotoxicity in North Indians. J Clin Pharm Ther 2014;39:91-6. [Crossref] [PubMed]
- Roy B, Ghosh SK, Sutradhar D, et al. Predisposition of antituberculosis drug induced hepatotoxicity by cytochrome P450 2E1 genotype and haplotype in pediatric patients. J Gastroenterol Hepatol 2006;21:784-6. [Crossref] [PubMed]
- Singla N, Gupta D, Birbian N, et al. Association of NAT2, GST and CYP2E1 polymorphisms and anti-tuberculosis drug-induced hepatotoxicity. Tuberculosis (Edinb) 2014;94:293-8. [Crossref] [PubMed]
- Sharma SK, Jha BK, Sharma A, et al. Genetic polymorphisms of CYP2E1 and GSTM1 loci and susceptibility to anti-tuberculosis drug-induced hepatotoxicity. Int J Tuberc Lung Dis 2014;18:588-93. [Crossref] [PubMed]
- Brito TC, Possuelo LG, Valim ARM, et al. Polymorphisms in CYP2E1, GSTM1 and GSTT1 and anti-tuberculosis drug-induced hepatotoxicity. An Acad Bras Cienc 2014;86:855-65. [Crossref] [PubMed]
- Teixeira RL, Morato RG, Cabello PH, et al. Genetic polymorphisms of NAT2, CYP2E1 and GST enzymes and the occurrence of antituberculosis drug-induced hepatitis in Brazilian TB patients. Mem Inst Oswaldo Cruz 2011;106:716-24. [Crossref] [PubMed]
- Santos NP, Callegari-Jacques SM, Ribeiro Dos Santos AK, et al. N-acetyl transferase 2 and cytochrome P450 2E1 genes and isoniazid-induced hepatotoxicity in Brazilian patients. Int J Tuberc Lung Dis 2013;17:499-504. [Crossref] [PubMed]
- Cho HJ, Koh WJ, Ryu YJ, et al. Genetic polymorphisms of NAT2 and CYP2E1 associated with antituberculosis drug-induced hepatotoxicity in Korean patients with pulmonary tuberculosis. Tuberculosis (Edinb) 2007;87:551-6. [Crossref] [PubMed]
- Kim SH, Kim SH, Bahn JW, et al. Genetic polymorphisms of drug-metabolizing enzymes and anti-TB drug-induced hepatitis. Pharmacogenomics 2009;10:1767-79. [Crossref] [PubMed]
- Chamorro JG, Castagnino JP, Musella RM, et al. Sex, ethnicity, and slow acetylator profile are the major causes of hepatotoxicity induced by antituberculosis drugs. J Gastroenterol Hepatol 2013;28:323-8. [Crossref] [PubMed]
- Vuilleumier N, Rossier MF, Chiappe A, et al. CYP2E1 genotype and isoniazid-induced hepatotoxicity in patients treated for latent tuberculosis. Eur J Clin Pharmacol 2006;62:423-9. [Crossref] [PubMed]
- Yamada S, Tang M, Richardson K, et al. Genetic variations of NAT2 and CYP2E1 and isoniazid hepatotoxicity in a diverse population. Pharmacogenomics 2009;10:1433-45. [Crossref] [PubMed]
- Artini IGA, Artana IGNB, Aman IGM, et al. CYP2E1 genotype and transaminase level of tuberculosis patients receiving fixed dose combination of antituberculosis. Bali Medical Journal 2017;6:S70-4. [Crossref]
- Sotsuka T, Sasaki Y, Hirai S, et al. Association of isoniazid-metabolizing enzyme genotypes and isoniazid-induced hepatotoxicity in tuberculosis patients. In Vivo 2011;25:803-12. [PubMed]
- Ben Fredj N, Gam R, Kerkni E, et al. Risk factors of isoniazid-induced hepatotoxicity in Tunisian tuberculosis patients. Pharmacogenomics J 2017;17:372-7. [Crossref] [PubMed]
- Sheng YJ, Wu G, He HY, et al. The association between CYP2E1 polymorphisms and hepatotoxicity due to anti-tuberculosis drugs: a meta-analysis. Infect Genet Evol 2014;24:34-40. [Crossref] [PubMed]
- Moreau A, Vilarem MJ, Maurel P, et al. Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol Pharm 2008;5:35-41. [Crossref] [PubMed]
- Zazuli Z, Barliana MI, Mulyani UA, et al. Polymorphism of PXR gene associated with the increased risk of drug-induced liver injury in Indonesian pulmonary tuberculosis patients. J Clin Pharm Ther 2015;40:680-4. [Crossref] [PubMed]
- Wang JY, Tsai CH, Lee YL, et al. Gender-dimorphic impact of PXR genotype and haplotype on hepatotoxicity during antituberculosis treatment. Medicine (Baltimore) 2015;94:e982 [Crossref] [PubMed]
- Wang Y, Xiang X, Huang WW, et al. Association of PXR and CAR polymorphisms and antituberculosis drug-induced hepatotoxicity. Sci Rep 2019;9:2217. [Crossref] [PubMed]
- Zhang J, Zhao Z, Bai H, et al. Genetic polymorphisms in PXR and NF-κB1 influence susceptibility to anti-tuberculosis drug-induced liver injury. PLoS One 2019;14:e0222033 [Crossref] [PubMed]
- Bao Y, Ma X, Rasmussen TP, et al. Genetic variations associated with anti-tuberculosis drug-induced liver injury. Curr Pharmacol Rep 2018;4:171-81. [Crossref] [PubMed]
- Huang JH, Zhang C, Zhang DG, et al. Rifampicin-induced hepatic lipid accumulation: association with up-regulation of peroxisome proliferator-activated receptor γ in mouse liver. PLoS One 2016;11:e0165787 [Crossref] [PubMed]
- Kim JH, Nam WS, Kim SJ, et al. Mechanism investigation of rifampicin-induced liver injury using comparative toxicoproteomics in mice. Int J Mol Sci 2017;18:1417. [Crossref] [PubMed]
- McDonagh EM, Boukouvala S, Aklillu E, et al. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics 2014;24:409-25. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


