
The value of gene chip detection of bronchoalveolar lavage fluid in the diagnosis of nontuberculous mycobacterial lung disease
Introduction
Nontuberculous mycobacteria (NTM) is widely present in natural environments such as water, soil, and dust, and most are parasitic bacteria. Only a small number of NTM cause human pathogenicity, and more than 190 types of NTM have been discovered to date (1). NTM can invade human lungs, lymph nodes, bones, joints, skin and soft tissues, and other tissues and organs, and can cause systemic disseminated diseases (2). With the gradual control of the tuberculosis epidemic and the development of strain identification techniques, NTM infection-associated diseases show a clear increasing trend and have become a public health issue that threatens human health (3). Lung disease due to NTM occurs commonly in structural lung disease, such as chronic obstructive pulmonary disease (COPD), bronchiectasis, cystic fibrosis (CF), pneumoconiosis, prior TB, pulmonary alveolar proteinosis, and esophageal motility disorders (4). Clinically, it is often difficult to differentially diagnose NTM and mycobacterium tuberculosis complex (MTBC)-induced mycobacteria lung disease, and these diseases are easily misdiagnosed. Therefore, accurate and rapid detection of mycobacteria and strain identification are of great importance for early diagnosis, timely and correct drug administration, and epidemiological surveillance of mycobacteria lung disease. At present, screening for NTM in Chinese laboratories generally involves isolating and culturing the mycobacterium first, and then the strains are identified. However, mycobacterial culture takes a considerable amount of time, and consequently, it is not possible to provide timely strain identification results for clinical use. Our research group aimed to directly examine the bronchoalveolar lavage fluid (BALF) of patients with NTM lung disease (NMLD) using gene chip technology (Capital Bio Corporation) and to compare the results with the sequencing results of MGIT960 cultured and isolated strains. We intended to investigate the value of the gene chip technique for the rapid diagnosis of NMLD using BALF.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/apm-21-1205).
Methods
Sample collection
BALF was collected from patients with suspected mycobacterial lung disease who underwent fiberoptic bronchoscopy at our hospital from January 2018 to June 2020. Both gene chip detection and BACTEC MGIT960 (Becton, Dickinson and Company) liquid mycobacterial culture were performed. A total of 308 specimens from patients with a diagnosis of NMLD were included in this study. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Fuzhou Institute of Tuberculosis Control and Prevention (No.:2019-001-01). Individual consent for this retrospective analysis was waived.
Main instruments and reagents
The mycobacterium culture apparatus MGIT960 (Becton, Dickinson and Company) and supporting reagents were purchased from BD Biosciences (USA). The Jingxing E-CyclerTM 96 PCR instrument, BioMixerTM II chip hybridization instrument, LuxScan10K/B microarray chip scanner, and mycobacterium strain identification chip were purchased from CapitalBio Corporation, China. The MPB64 antigen detection kit was purchased from Genesis Biodetection and Biocontrol Co., Ltd., Hangzhou, China. We used the 16S-23S rRNA intergenic transcribed spacer (ITS) and 16S rDNA homologous sequence analysis to identify the NTM strain from the isolated strains. Sequencing was performed by Sangon Biotech Company, Shanghai, China.
Isolation, culture and preliminary identification of mycobacterium by BACREC MGIT960
Mycobacterium were cultured according to the MGIT960 manual (Becton, Dickinson and Company). The cultured positive strains were identified as acid-fast bacteria positive by acid-fast staining. The MPB64 antigen detection method (Genesis Corporation, Hangzhou, China) was used for the initial identification of acid-fast strains: MPB64 antigen positive strains were MTBC, and negative strains were NTM.
Identification of bacterial strains in BALF by gene chip detection
Nucleic acid extraction
Two milliliters of BALF was added to an equal amount of 10% NaOH and mixed by shaking. After standing still for liquidation for 15 min, the mixture was centrifuged at 12,000 rpm/min for 2 min, and the supernatant was discarded. One milliliter of wash solution was added to the centrifuge tube, vortexed, and mixed. After centrifugation at 12,000 rpm for 2 min, the supernatant was discarded; 50 μL of nucleic acid extraction solution was added and mixed well and transferred to a nucleic acid extraction tube, which was placed in a nucleic acid extraction machine for 5 min with shaking, followed by a 95 °C water bath for 5 min and centrifugation at 10,000 rpm for 1 min. The supernatant was collected for nucleic acid detection.
PCR amplification
PCR was performed in strict accordance with the manual for bacterial strain identification by gene chip (CapitalBio Technology). The amplification reaction system was 20 μL, which included 18 µL for the amplification reaction solution and 2 µL for the DNA template. Pre-denaturation took place at 94 °C for 10 min; after a hot start, the solution was subjected to 45 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s, then 20 cycles at 94 °C for 30 s and 72 °C for 60 s and then kept at 72 °C for 7 min.
Hybridization and determination of the results
The PCR products were denatured in a PCR machine at 95 °C for 5 min, followed by an ice bath for 5 min. A hybridization mixture was prepared at a ratio of 9 μL of hybridization buffer and 6 μL of PCR products. A total of 13.5 μL of hybridization mixture was added to the chip array through the wells, and the loaded chips were placed in the BioMixerTM II microarray hybridization station (CapitalBio Corporation). Hybridization was carried out at 50 °C for 2 h, and the rotation speed was 5 rpm. After hybridization, the microarrays were washed and dried, and the signals were read and the results were determined using a LuxScanTM 10KB microarray chip scanner and a Jingxing mycobacterial strain identification chip discrimination system (CapitalBio Corporation).
Microbial strain identification of isolated strains
Nucleic acid extraction of strains
The nucleic acids of the strains were extracted by boiling. After mixing in mycobacterium culture-positive MGIT tubes (Becton, Dickinson and Company), 0.5 mL of bacterial solution was aspirated and placed in a water bath, boiled and lysed at 100 °C for 30 min, and centrifuged at 12,000 rpm for 10 min. The supernatant was used for the detection of nucleic acids.
PCR amplification and sequencing
In accordance with the literature, ITS homologous sequence analysis was used, and 16S rDNA sequencing was used for bacteria that could not be distinguished by using this method.
The ITS upstream primer was 5'-CGACGAAGTCGTAACAAGG-3', and the downstream primer was 5'-GCGTATCCNTTAGATGCTC-3'. The 16S rDNA upstream primer was 5'-CACATGCAAGTCGAACGGAAAGG-3'; the downstream primer was: 5'-GCCCGTATCGCCCGCACCCT-3'. The primers were synthesized by Sangon Biotech Company. The amplification reaction system comprised 25 µL, including 20 µL of amplification reaction solution and 5 µL of DNA template. The PCR amplification program was as follows: pre-denaturation at 95 °C for 5 min; after a hot start, the solution was subjected to 55 cycles at 95 °C for 15 s, 54 °C for 20 s, and 72 °C for 20 s, and then kept at 72 °C for 5 min. The amplified products were analyzed by agarose gel electrophoresis and sent to Sangon Biotech Company. The gene sequences were compared with the GeneBank Database of the National Center for Biotechnology Information (https:/blast.ncbi.nlm.nih.gov/B1ast.cgi) for homology analysis to identify the strains.
Diagnostic criteria for NMLD
Diagnostic criteria for NMLD based on the ATS/ERS/ESCMID/IDSA Clinical Practice Guideline for NTM (5).
Statistical analysis
SPSS 22.0 software was used for the statistical analysis. The chi-square test was used to compare count data among groups. P˂0.05 was considered statistically significant.
Results
Age and sex distribution of NTM patients
Among the 308 NMLD patients, 202 (65.57%) were female and 106 (34.43%) were male; the male-to-female ratio was 1:1.91. Females outnumbered males in all but the under 30 years age group. The distribution of NMLD in all age groups showed that middle-aged and elderly patients (i.e., those over 40 years of age) were the main population; these age groups accounted for 92.21% of the cases (284/308), and the 51 to 60 years age group was the largest (35.72%). The specific distribution characteristics are shown in Table 1.
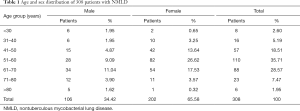
Full table
Comparison of test results between the gene chip and liquid culture methods
Among the 308 NMLD patients, 221 had positive BALF results according to the gene chip assay, for a positive rate of 71.75% (221/308); in the preliminary strain identification, the number of cases with positive NTM according to the liquid culture and MPB64 antigen assay was 218, for a positive rate of 70.78% (218/308). The gene chip method and the liquid culture method had a positive coincidence rate of 85.78%, a negative coincidence rate of 62.2%, and an overall coincidence rate of 78.90%, and there was no difference in the positive detection rate of NTM in BALF specimens between the two methods (χ2=0.138, P=0.804>0.05) (Table 2). There were 218 cases of positive NTM in liquid cultures of BALF; they were reported at a range of 5 to 36 days, with a mean time of 21.49 days and a median time of 21 days; in comparison, the entire gene chip detection process took 6 h.
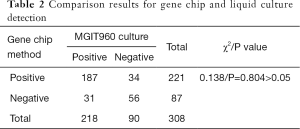
Full table
Comparison of NTM strain identification using the gene chip method and gene sequencing technology
A total of 218 strains of NTM were isolated from the BALF specimens of 308 NMLD patients after being cultured in liquid. After nucleic acid extraction, PCR amplification was performed, followed by ITS and 16S rDNA sequencing. Eight species were identified among the 218 NTM strains, including M. intracellulare in 131 strains (60.09%), M. avium in 48 strains (22.02%), M. abscessus in 27 strains (12.38%), M. kansasii in five strains (2.29%), M. szulgai/malmoense in two strains (0.92%), and one strain each of M. scrofulaceum, M. marseillense, and M. senegalense (0.46%). The gene chip method was used to directly detect NTM in BALF. A total of 221 cases of NTM were detected in 308 samples, and 6 species were identified. The order of strain distribution was the same as that of the sequencing results (Table 3).
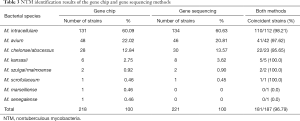
Full table
Among the 308 BALF specimens, 187 were simultaneously analyzed by gene sequencing and gene chip detection, and a total of 181 cases had coincident strain identification results. The coincidence rate of the two methods was 96.79%. The gene chip and gene sequencing results were inconsistent in a total of 6 cases (Table 4). In addition, because gene chip technology cannot distinguish M. chelonae from M. abscessus or M. szulgai from M. malmoense, the detection results were represented as M. chelonae/abscessus and M. szulgai/malmoense. The sequencing results of the 28 cases of M. chelonae/abscessus were all M. abscessus. The sequencing results of the two cases of M. szulgai/malmoense were all M. malmoense.
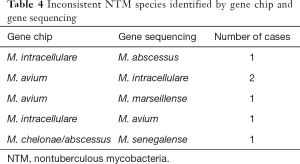
Full table
Discussion
In recent years, NTM has become an important pathogen that threatens human health, and the incidence and prevalence of NMLD continue to rise worldwide (6). A retrospective study in the USA showed that the incidence of NMLD increased from 3.13/100,000 in 2008 to 11.70/100,000 in 2015 (7). The infection rate of NTM has also shown a trend toward rapid growth in China. An epidemiological survey showed that the rate of NTM isolation in clinically isolated mycobacteria increased from 11.1% to 22.9% over a decade (8). NTM can infect multiple tissues and organs in the human body, but the lungs are the most common infection site, accounting for approximately 65–90% of cases (9). The symptoms, signs, and imaging findings of NMLD are very similar to those of pulmonary tuberculosis. However, NTM is generally resistant to common anti-TB drugs. Therefore, the rapid detection and identification of NTM species is very important for the diagnosis and treatment of NMLD. At present, most of the reported NTM strain identification methods are based on cultures to obtain isolates before strain identification (10-12). Due to the long cultivation time of mycobacteria, it takes 1–8 weeks and sometimes as long as 12 weeks to obtain a culture (13). Therefore, this method cannot provide timely and rapid clinical information. CapitalBio GeneChip technology is a commercially available indirect homologous sequence comparison method that uses 16S rDNA-specific single-nucleotide polymorphic sites as probes and fixes the probes on a chip after labeling. It indirectly determines the DNA sequence composition through the binding of the probes to the sequences to be tested, thus providing strain identification. This method is simple and can be performed quickly; the whole process takes only 6 h. It can identify 17 clinically common NTMs, including M. tuberculosis, M. intracellulare, M. chelonae/M. abscessus, M. kansasii, M. avium, M. gordonae, M. fortuitum, M. scrofulaceum, M. gilvum, M. terrae, M. phlei, M. nonchromogenicum, M. marinum/M. ulcerans, M. aurum, M. szulgai/M. malmoense, M. xenopi, and M. smegmatis. Reliable results have been reported in the literature for the identification of isolated strains and sputum specimens (14,15). Although molecular biology technology has shown high sensitivity for the detection of MTBC in BALF (16,17), there is no report on the use of BALF specimens for NTM detection; therefore, this study intended to investigate the diagnostic value of gene chip technology for NMLD by statistically analyzing the results of gene chip detection of NTM in BALF from patients with NMLD with the goal of aiding in the rapid diagnosis and treatment of NMLD and providing additionally clinical data on the prevalence of NTM in Fujian Province to guide the prevention and treatment of NMLD.
In this study, a total of 308 BALF specimens underwent simultaneous detection using the liquid culture method and the gene chip method, and the culture method was used as the reference method for evaluating the NTM detection rate of the gene chip method. The results showed that 221 out of 308 BALF samples from NMLD patients had positive results with gene chip detection, with a positive rate of 71.75% (221/308), while the number of positive cases detected with the liquid culture method and preliminarily identified as NTM was 208, with a positive rate of 70.78% (218/308); there was no difference in the positive detection rate of NTM in BALF between the two methods (χ2=0.138, P>0.05). Compared to the liquid culture method, which requires an average of 21 days to detect NTM, the entire gene chip detection process takes only 6 h, which greatly decreases the detection time and allows clinicians to quickly diagnose NMLD.
In this study, the liquid culture-positive strains were sequenced using ITS and 16S rDNA gene sequencing and compared with the strain identification results directly detected in the BALF using the gene chip method to evaluate the accuracy of strain identification by gene chip. A total of 187 samples were subjected to simultaneous gene sequencing and direct gene chip detection of the isolated strains. A total of 181 strains had matching strain identification results; the coincidence rate between the two results was 96.79%, which was very high. A multicenter study in China in which the gene chip method was used for direct detection in positive sputum specimens also showed a fairly high coincidence rate between the gene chip method and the gene sequencing method (15), and coincidence rate of NTM strain identification and the sequencing of sputum culture isolates reached 95.4%. This study showed that gene chip technology can accurately identify NTM strains in BALF without the need for strain isolation and culturing, which decreases both the time and cost of detection. However, this technique has some limitations. First, this technique is based on the detection of 16S rDNA single sequences; even though this sequence currently has the most complete database for strain identification, its similarity among NTM strains is as high as 90%, its differentiation ability is low, and it cannot accurately distinguish M. chelonae from M. abscessus, M. szulgai from M. malmoense, or M. marinum from M. ulcerans (18). Therefore, ITS was selected as the main sequence for the sequencing method to improve the ability to differentiate among strains. Second, only 17 common clinical NTMs can be identified, which a small number compared to the large variety of pathogenic NTMs. In this study, M. marseillense and M. senegalense were not included in the detection range and were incorrectly identified. In view of these limitations, we suggest the use of a high-throughput gene chip, such as the ITS sequence, to increase the probe sequence and the probe types of pathogenic NTM to improve the differentiation ability and the number of strains identified.
The majority of the 308 NMLD patients included in this study were female and middle-aged or elderly; the number of female patients was 1.91 times that of male patients, and the proportion of menopausal women over 50 years of age was 48.05% (148/308), which is basically consistent with the literature reports (19). The sequencing results for 218 strains of NTM isolate showed that the main pathogens of NMLD in our hospital were M. intracellulare, M. avium, M. abscessus, and M. kansasii, accounting for as much as 97.71% (213/218). These four types of NTM are also the most common strains of NMLD reported domestically and abroad (5,20,21). Among them, M. intracellulare and M. avium are collectively referred to as MAC. This group represents the mycobacteria for which the highest number of new strains or subspecies are found and comprises the main pathogen of NMLD at our hospital. In our study, 3 species were found: M. intracellulare in 131 strains, M. avium in 48 strains, and M. marseillense in 1 strain; together, they accounted for up to 82.57% of the isolated strains (180/218). Although the prevalence of NTM shows significant geographical variation, overall, the isolation rate of MAC is the highest, and MAC lung disease is the most common NMLD observed (22). This study shows that the coincidence rate of gene chip technology and sequencing for the detection of the most common strains of NMLD—MAC, M. abscessus, and M. kansasii—were 97.42% (151/155), 95.65% (41/42), and 100% (6/6), respectively. Gene chip detection in BALF is sufficient for the preliminary clinical diagnosis of NTM.
In summary, the incidence of NMLD shows an increasing yearly trend, and it is easily confused with tuberculosis (TB). NTM has high resistance to most anti-TB drugs; therefore, the rapid detection and identification of NTM at the species level is of great clinical importance. Gene chip technology is a molecular biology technology developed in recent years. It has the characteristics of rapidness, high throughput, and a high degree of automation. The detection of NTM in BALF using this technology can help clinicians rapidly diagnose NMLD. Additionally, it enables clinicians to select suitable drug sensitivity tests in a timely manner based on the provided information about the bacterial strains, which allows a shift from empirical drugs to targeted drugs and provides a basis for precise clinical treatment.
Acknowledgments
Funding: Fujian Provincial Health Technology Project for the Young and Middle-aged Backbone Talent Training Program (2019-ZQN-89), Fuzhou Science and Technology Project (2019-SZ-59).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-1205
Data Sharing Statement: Available at https://dx.doi.org/10.21037/apm-21-1205
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-1205). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Fuzhou Institute of Tuberculosis Control and Prevention (No.:2019-001-01). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kong L, Xie B, Liu Q, et al. Application of acid-fast staining combined with GeneXpert MTB/RIF in the diagnosis of non-tuberculous mycobacteria pulmonary disease. Int J Infect Dis 2021;104:711-7. [Crossref] [PubMed]
- Henkle E, Hedberg K, Schafer SD, et al. Surveillance of Extrapulmonary Nontuberculous Mycobacteria Infections, Oregon, USA, 2007-2012. Emerg Infect Dis 2017;23:1627-30. [Crossref] [PubMed]
- Shih DC, Cassidy PM, Perkins KM, et al. Extrapulmonary Nontuberculous Mycobacterial Disease Surveillance - Oregon, 2014-2016. MMWR Morb Mortal Wkly Rep 2018;67:854-7. [Crossref] [PubMed]
- Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 2007;175:367-416. Erratum in: Am J Respir Crit Care Med 2007;175:744-5. [Crossref] [PubMed]
- Daley CL, Iaccarino JM, Lange C, et al. Treatment of Nontuberculous Mycobacterial Pulmonary Disease: An Official ATS/ERS/ESCMID/IDSA Clinical Practice Guideline. Clin Infect Dis 2020;71:905-13. [Crossref] [PubMed]
- Stout JE, Koh WJ, Yew WW. Update on pulmonary disease due to nontuberculous mycobacteria. Int J Infect Dis 2016;45:123-34. [Crossref] [PubMed]
- Winthrop KL, Marras TK, Adjemian J, et al. Incidence and Prevalence of Nontuberculous Mycobacterial Lung Disease in a Large U.S. Managed Care Health Plan, 2008-2015. Ann Am Thorac Soc 2020;17:178-85. [Crossref] [PubMed]
- Zhang Z, Pang Y, Wang Y, et al. Differences in risk factors and drug susceptibility between Mycobacterium avium and Mycobacterium intracellulare lung diseases in China. Int J Antimicrob Agents 2015;45:491-5. [Crossref] [PubMed]
- Porvaznik I, Solovič I, Mokrý J. Non-Tuberculous Mycobacteria: Classification, Diagnostics, and Therapy. Adv Exp Med Biol 2017;944:19-25. [Crossref] [PubMed]
- Chae H, Han SJ, Kim SY, et al. Development of a One-Step Multiplex PCR Assay for Differential Detection of Major Mycobacterium Species. J Clin Microbiol 2017;55:2736-51. [Crossref] [PubMed]
- Chikamatsu K, Aono A, Kawai A, et al. Evaluation of Q Gene Mycobacteria: A novel and easy nucleic acid chromatography method for mycobacterial species identification. J Microbiol Methods 2019;163:105657 [Crossref] [PubMed]
- Rodriguez-Temporal D, Rodríguez-Sánchez B, Alcaide F. Evaluation of MALDI Biotyper Interpretation Criteria for Accurate Identification of Nontuberculous Mycobacteria. J Clin Microbiol 2020;58:e01103-20. [Crossref] [PubMed]
- Brown-Elliott BA, Nash KA, Wallace RJ. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 2012;25:545-82. [Crossref] [PubMed]
- Zhu L, Jiang G, Wang S, et al. Biochip system for rapid and accurate identification of mycobacterial species from isolates and sputum. J Clin Microbiol 2010;48:3654-60. [Crossref] [PubMed]
- Fang H, Shangguan Y, Wang H, et al. Multicenter evaluation of the biochip assay for rapid detection of mycobacterial isolates in smear-positive specimens. Int J Infect Dis 2019;81:46-51. [Crossref] [PubMed]
- Pan X, Yang S, Deighton MA, et al. A comprehensive evaluation of Xpert MTB/RIF assay with bronchoalveolar lavage fluid as a single test or combined with conventional assays for diagnosis of pulmonary tuberculosis in China: a two-center prospective study. Front Microbiol 2018;9:444. [Crossref] [PubMed]
- Le Palud P, Cattoir V, Malbruny B, et al. Retrospective observational study of diagnostic accuracy of the Xpert® MTB/RIF assay on fiberoptic bronchoscopy sampling for early diagnosis of smear-negative or sputum-scarce patients with suspected tuberculosis. BMC Pulm Med 2014;14:137. [Crossref] [PubMed]
- Ninet B, Monod M, Emler S, et al. Two Different 16S rRNA Genes in a Mycobacterial Strain. J Clin Microbiol 1996;34:2531-6. [Crossref] [PubMed]
- Kwon YS, Koh WJ. Diagnosis and Treatment of Nontuberculous Mycobacterial Lung Disease. J Korean Med Sci 2016;31:649-59. [Crossref] [PubMed]
- Zhou L, Xu D, Liu H, et al. Trends in the Prevalence and Antibiotic Resistance of Non-tuberculous Mycobacteria in Mainland China, 2000-2019: Systematic Review and Meta-Analysis. Front Public Health 2020;8:295. [Crossref] [PubMed]
- Singh K, Kumari R, Tripathi R, et al. Detection of clinically important nontuberculous mycobacteria (NTM) from pulmonary samples through one-step multiplex PCR assay. BMC Microbiol 2020;20:267. [Crossref] [PubMed]
- Larsson LO, Polverino E, Hoefsloot W, et al. Pulmonary disease by non-tuberculous mycobacteria - clinical management, unmet needs and future perspectives. Expert Rev Respir Med 2017;11:977-89. [Crossref] [PubMed]


