Rapid detection of multiple pathogens by the combined loop-mediated isothermal amplification technology and microfluidic chip technology
Introduction
With the wide application of antibiotics and the increase of invasive operations, patients, especially a large number of critically ill patients, have become high-risk groups for a variety of pathogenic bacterial infections due to their critical condition, low immunity, multiple organ failure, frequent invasive operations and exposure to a variety of antibiotics. After infection, most patients will be more serious, the cost of treatment will increase, and even endanger the lives of patients. Therefore, timely and accurate detection of pathogenic bacteria is not only the basis for the diagnosis of infectious diseases, but also the basis for selecting individualized treatment protocols and standardizing the use of antibiotics. However, it is difficult to diagnose infectious diseases accurately in clinical practice because hair coloring pathogens are various, and patients may carry more than one pathogen. They often coexist when infection occurs, and which makes it difficult to diagnose infectious diseases. Therefore, improved awareness of multiple bacterial infections and the ability to determine the pathogen as soon as possible are urgently needed. Traditional bacterial culture methods are the gold standard for identifying pathogens of infectious diseases. Most current laboratories currently continue to use traditional culture and detection methods. The culture method involves complicated procedures, high requirements for operators, cross-interference and considerable time cost However, the, accurate, timely, and rapid identification of pathogen types is extremely important for effective treatment of infectious diseases or incidents. Furthermore, the harsher environment typical of immediate inspection conditions, such as public health infection incident sites, postdisaster treatment sites, and small, grassroots hospitals place even greater challenges on the accuracy and effectiveness of the inspection methods. Loop-mediated Isothermal nucleic acid amplification (LAMP) is a single tube technique for the amplification of DNA. It provides a low cost alternative to polymerase chain reaction (PCR) technology to detect certain diseases. Studies have shown LAMP can be a rapid diagnostic for COVID-19. The microfluidic chip is an attractive miniaturized platform with valuable advantages such as low cost analysis, low reagent consumption, reduced sample volume, and shortened processing time. In this study, LAMP technology and microfluidic chips were integrated and optimized to establish simultaneous detection of multiple pathogens, and multiple detection and real-time detection were realized in a closed system without aerosol contamination within 40 minutes.
We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/apm-21-2792).
Methods
Research object
Experimental strains
Escherichia coli [American Type Culture Collection (ATCC)25922], Acinetobacter baumannii (ATCC BAA-747), Staphylococcus aureus (ATCC29213), Stenotrophomonas maltophilia (ATCC17666), Enterococcus faecalis (ATCC29212), Carbapenem-resistant enterobacteriaceae (CRE), Carbapenem-resistant Pseudomonas aeruginosa (CRPA), Methicillin-resistant Staphylococcus aureus (MRSA), Methicillin-resistant Coagulase Negative Staphylococcus (MRCNS), Burkholderia cepacia, Pneumoniae klebsiella, Shigella, Staphylococcus epidermidis, Enterococcus faecium, and Candida albicans were all preserved by the center of Medical School of Chinese PLA. Good resuscitation and cultivated activity of bacteria was confirmed by the bacterial identification, drug susceptibility system, and microbial identification mass spectrometry.
Instruments and reagents
This study acquired the following instruments and reagents: a VITEK MS microbial identification mass spectrometer (bioMérieux SA, Marcy L’Etoile, France), a Nano-400A micro spectrophotometer (Aosheng Instrument, Hangzhou, China), a high-speed tissue grinder KZ-11 (Servicebio Technology. Wuhan, China), a metal bath (Kylin-Bell, Haimen, China), a BacT/ALERT 3D automatic culture system and VITEK-2 compact ID/AST (bioMérieux SA, Marcy L'Etoile, France), a blood agar plate (Zhengzhou Antu Bioengineering Co., Ltd. Zhengzhou, China), DNA lamp amplification reaction kits and a microfluidic chip detector (Biocare, Tianjin, China), and a bacterial genome extraction kit and lysozyme (Tiangen Biotechnology, Beijing, China). The primers were synthesized by Beijing Synthesis Department of Biotechnology (Sangon Biotech, Shanghai, China).
Experimental method
Design and synthesis of primers
Strains and nucleic acid sequences of multiple target fragments were obtained from the Nucleotide database on the National Center for Biotechnology Information (NCBI) official website. The conserved regions were screened in sequence alignment software, and the conserved regions of target sequences less than 2000 bp were selected as the design template. Target conservative sequences for primer design were uploaded to the LAMP online primer design website (http://primerexplorer.jp/lampv5e/index.html). Sequence alignment of the functional area was conducted using the Basic Local Alignment Search Tool (BLAST) on the NCBI official website to evaluate the specificity and conservation of the primers. After the primers were designed and synthesized, multiple sets of primers were screened to select the best primers. LAMP analysis of each strain involved 4 to 6 primers, including 2 external primers (F3 and B3), 2 internal primers (FIP and BIP), and loop primers (LF and/or LB). Loop primers were not necessary for LAMP reactions. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Extraction of bacterial genomic DNA
The extraction of experimental strains and sample DNA was performed in accordance with the instructions of Biocare bacteria and sample genomic DNA extraction kit. Through the preliminary pretreatment, 200 µL of the pretreated bacterial suspension was centrifuged at a speed of 12,000 rpm for 2 minutes, and the supernatant was discarded. Then, the pellet was resuspended in 200 µL of extraction solution containing 1 mM ethylenediamine tetra-acetic acid, 10 mM of Tris-HCl, and nucleic acid protector, and then it was transferred to a lysis tube containing glass beads. The tube was vibrated for 5 minutes by vortex mixer. Afterwards, it was heated at 100 °C for 5 minutes. The centrifuge tubes were briefly centrifuged to obtain the supernatant. For the positive culture of the lower respiratory tract secretion sample, trypsin was added into the sample, and it was mixed by vortex thoroughly, and liquefied at 37 °C for 30 minutes until it was completely liquefied without any solid and stringy material, whereupon DNA extraction was performed. The concentration and purity of the extracted DNA were detected, and the DNA was stored at –20 °C for further application.
Reaction of DNA-LAMP detection
The constant-temperature amplification reagent was taken out from the kit, fully dissolved, vibrated and mixed by a vortex mixer, and centrifuged instantaneously. In the reagent storage and preparation area, 58 µL of constant-temperature amplification reagent was added to the prepared 200 µL centrifuge tube, the cap was closed, and the solute was moved to the specimen preparation area. Each sample was placed into a single centrifuge tube. In the specimen preparation area, 14.5 µL of nucleic acid was added to be tested, vibrated, and mixed well by vortex, and then immediately centrifuged. Each nucleic acid amplification reaction system was 72.5 µL. In the specimen preparation area, the chip was taken out from the kit and kept at room temperature; the chip was unpacked on a clean workbench and placed with the inlet upwards. Next, 70 µL of reaction system of the prepared nucleic acid amplification was sucked up by a pipette and added to the main channel of the chip through the sample inlet. When it fills the entire chip channel, and you can feel the obvious resistance to adding the sample, stop adding the sample. Then press the buckles of the injection hole and the vent hole respectively. The sample amount of the reaction system is shown in Table 1. The cutoff turbidity value was fixed at 0.1 to differentiate positive from negative results.
Table 1
| Component of reaction | Volume (μL) |
|---|---|
| Isothermal amplification reagent | 58.0 |
| Template DNA solution | 14.5 |
| Total | 72.5 |
Design and processing of microfluidic chips
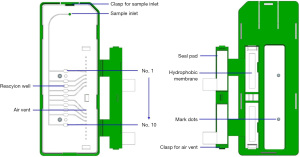
The microfluidic chip used in this study was designed by SolidWorks 2014 software, and injection molding of medical grade polycarbonate (PC) was implemented. The chip contained 10 independent reaction chambers, each with a volume of about 4 µL, and the reaction chambers was connected by a slender 500 µM × 200 µM channel. The chip structure is displayed in Figure 1. When the chip was assembled, the injection-molded PC layer first was cleaned with ethanol and pure water by an ultrasonic cleaning machine for 2 minutes, and then it was dried in an oven at 80 °C for 3 hours. The various primers listed in Table 1 were put into the corresponding reaction chambers. Each chip was equipped with 3 quality-control reaction wells, including 1 AC (active control, AC) for the sample of yeast DNA and corresponding primers. The entire quality of the experimental reaction process was monitored to monitor whether the sample collection was qualified, 1 IC internal quality control was used for the sampling point of a section of human genome DNA and corresponding primers, and 1 BC (blank control, BC) was designed without any primers. The effectiveness of each test was monitored by a combination of 3 quality control modes. After sample loading, the solution was dried at room temperature for 1 hour. Finally, the chip was sealed with tape and stored at 0–4 °C for later application, and then the samples were added until equilibrated to room temperature during use.
Microfluidic chip detector
The microfluidic chip detector is mainly composed of a temperature control module and a detection module, with a volume of 35 cm × 34 cm ×18 cm and a weight of about 8 kg, as shown in Figure 2. The temperature control module could provide a Isothermal of 65 °C for constant-temperature amplification. The EvaGreen dye detection module was excited by blue LEDs (light-emitting diode) and optical filters, and a high-definition camera recorded the fluorescence of the reaction chamber for detection. During the LAMP reaction, the intensity was calculated by OpenCV according to the fluorescence intensity, and a real-time curve was drawn and displayed on the screen of the detector. The instrument can process 4 microfluidic chips in batches synchronously.
Evaluation of results
In the constant-temperature amplification reaction system, the real-time fluorescence detection was carried out by adding fluorescent dyes using the fluorescent dye incorporation method. An “S”-shaped real-time fluorescence amplification curve was generated when the positive sample was amplified and detected. A positive quality control test result should be positive, while a negative quality control test result should negative, which would indicate a valid test result. If the quality control result was incorrect, this was judged as an invalid test result, which would require re-inspection or find the reason.
Statistical analysis
The data were calculated using the descriptive research method. The experimental data were converted into the 4-grid table format shown in Table 2 and calculated according to the following formulae: positive coincidence rate = a/(a + c)×100%; negative coincidence rate = d/(b + d)×100%; total coincidence rate = (a + d)/(a + b + c + d)×100%. The diagnostic performance of the chip was evaluated by receiver operating characteristic (ROC) curve.
Table 2
| Positive (reference method) | Negative (reference method) | Total | |
|---|---|---|---|
| Positive (experimental method) | a | b | a+b |
| Negative (experimental method) | c | d | c+d |
| Total | a+c | b+d | a+b+c+d |
Results
Screening of LAMP primers
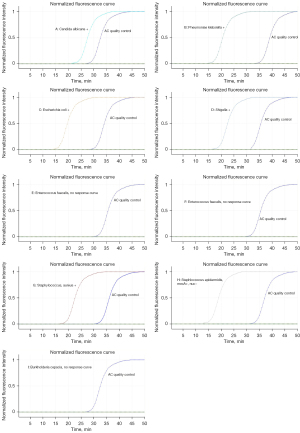
Primers were designed and screened for the conservative genes of the pathogen and amplified by LAMP method. The positive reaction showed a typical S-shaped curve, and the negative control had no S-shaped curve. The first S-shaped curve was the best primer, and the best primer sequence is shown in Table 3.
Table 3
| Name of bacteria | Target gene | Primer name | Primer sequence |
|---|---|---|---|
| Escherichia coli | uidA | F3 | GCATCGTGGTGATTGAATG |
| B3 | GTTCGTTGGCAATACTGCCC | ||
| FIP | TCTTTCGGCTTGTTGCCCGCCTGCTGTCGGCTTTAACCTC | ||
| BIP | TACAGCGAAGAGGCAGTCAACGTTTTGGTTTTTGTCACGCGCTATC | ||
| LF | CGAAACCAATGCCTGAGGT | ||
| LB | GCACTTACAGGCGAAA | ||
| Shigella | ipaH | F3 | GCAGATTTACTTCTCCATGAGT |
| B3 | GTATTGCGTGCAGAGACG | ||
| FIP | GTTTGTTTTCCGGGAACCATGC-GACGGACAACAGAATACACT | ||
| BIP | GATGTATCACAGATATGGCATGCT-GAAAGGCGGTCAAGGAAC | ||
| LF | ATCAGCCAGGGGGCGAT | ||
| MRSA | mecA | F3 | TGATGCTAAAGTTCAAAAGAGT |
| B3 | GTAATCTGGAACTTGTTGAGC | ||
| FIP | TGAAGGTGTGCTTACAAGTGCTAAT-CAACATGAAAAATGATTATGGCTC | ||
| BIP | TGACGTCTATCCATTTATGTATGGC-AGGTTCTTTTTTATCTTCGGTTA | ||
| LF | TCACCTGTTTGAGGGTGGA | ||
| Staphylococcus aureus | Nuc | F3 | AACAGTATATAGTGCAACTTCAA |
| B3 | CTTTGTCAAACTCGACTTCAA | ||
| FIP | ATGTCATTGGTTGACCTTTGTACAT-AAATTACATAAAGAACCTGCGA | ||
| BIP | GTTGATACACCTGAAACAAAGCATC-ATTTTTTTCGTAAATGCACTTGC | ||
| LF | GTATCACCATCAATCGCTTT | ||
| LB | GGTGTAGAGAAATATGGTCC | ||
| Pneumoniae klebsiella | fimD | F3 | TCTCGCCAAGCGGTCC |
| B3 | TGAGACCTACTGGGTGT | ||
| FIP | AACCGCAATGTGCAAAGCAGTCAGGGAGACGACCGT | ||
| BIP | AGTTAATAAAGTAGGAGGCCTTGCTTTTGTTCGCGTATGGAATCGG | ||
| LF | ATGACAGCAGCAATAAGGC | ||
| LB | CTATAGCCCACGCGGT | ||
| Candida albicans | IFA22 | F3 | CCCTGTATGGATATCATTCTCTG |
| B3 | AGTGGAGTGATGAACCAG | ||
| FIP | AAGACTATTTGGGATACTTTCCCGCTCCTCCCACAAAAAGAAACC | ||
| BIP | GGTACACATGCCATTGAGTTATGCTAAATGCGACGTGTTCGA | ||
| LF | AATGCTCTGAGCATTAAG | ||
| LB | CCTCCTTAAAATCACTGAA |
MRSA, methicillin-resistant
Determination of the positive cutoff value
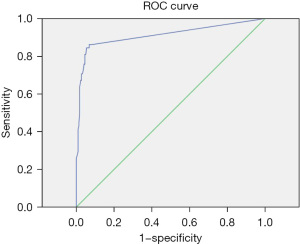
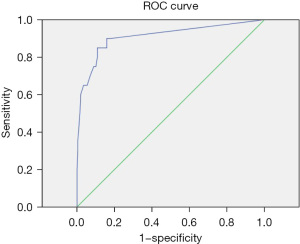
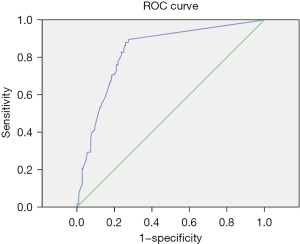
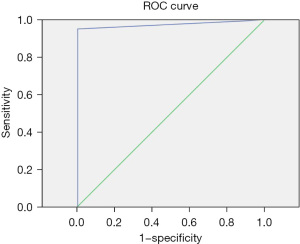
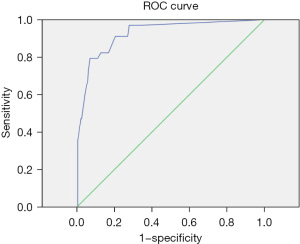
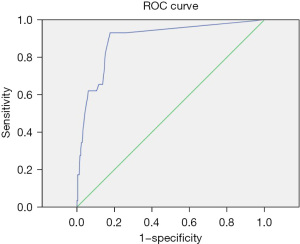
The positive judgment value of 6 kinds of pathogens was determined by ROC curve analysis. The ROC curve was drawn, the best cutoff values for Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, MRSA, and Candida albicans were 38.5, 21.25, 31.5, 36.5, 22.5, and 33.75, respectively. The sensitivities corresponding to the cutoff value were 86%, 85%, 88%, 95%, 97%, and 93%, respectively. The specificity was 93%, 89%, 74%, 100%, 90%, and 82%, respectively. The area under the curve was 0.91, 0.91, 0.83, 0.97, 0.96, and 0.9, respectively. All showed good diagnostic performance, and the results are shown in Table 4 and Figures 3-8.
Table 4
| Name of bacteria | Best cutoff value | Sensitivity | Specificity | Area under the curve | Confidence interval |
|---|---|---|---|---|---|
| Staphylococcus aureus | 38.5 | 0.86 | 0.93 | 0.91 | 0.86–0.97 |
| Escherichia Coli | 21.25 | 0.85 | 0.89 | 0.91 | 0.83–0.99 |
| Pneumoniae klebsiella | 31.5 | 0.88 | 0.74 | 0.83 | 0.77–0.89 |
| Shigella | 36.5 | 0.95 | 1 | 0.97 | 0.92–1 |
| MRSA | 22.5 | 0.97 | 0.9 | 0.96 | 0.92–1 |
| Candida albicans | 33.75 | 0.93 | 0.82 | 0.9 | 0.83–0.96 |
MRSA, methicillin-resistant
Detection limit of microfluidic chip
After adjusting the strain to a 0.8–0.9 McDonald's bacterial suspension with a McDonald's turbidity meter, the DNA of bacteria was extracted. The concentration and purity of the extracted DNA of Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, and MRSA were detected. After 10-time gradient dilution with sterile ddH2O, the solution was added into the microfluidic chip and detected by the microfluidic detection system, with the lowest detectable DNA concentration being considered as the detection limit of the detection system. The dilution multiples were 10, 102, 103, 104, 105, and 106 times. The detection limit of each bacterial type is shown in Table 5.
Table 5
| Name of bacteria | Dilution factor for positive results | Detection limit (pg/μL) |
|---|---|---|
| Staphylococcus aureus | 103 | 6.95 |
| Escherichia Coli | 104 | 44.6 |
| Pneumoniae klebsiella | 104 | 3.89 |
| Shigella | 104 | 15.33 |
| MRSA | 103 | 16.45 |
| Candida albicans | 102 | 463 |
MRSA, methicillin-resistant
Anti-interference ability of microfluidic chip
DNA extraction was performed on 15 kinds of bacteria identified by a microbial mass spectrometer and microbial identification instrument. Then the routine detection was performed to observe the influence of the DNA of other bacterial species that may have cross-reactions on the detection accuracy. The experimental results from the 15 species of bacteria were positive for the target bacteria, while the results of the other species were negative, indicating that the detection of target bacteria by the microfluidic detection system did not cross-react with the detection of the other 15 strains by this method, as shown in Table 6 and Figure 9.
Table 6
| No. | Type | Target gene | Negative and positive |
|---|---|---|---|
| 1 | Staphylococcus, aureus (Sa) | nuc | nuc + |
| 2 | Escherichia coli (E coli) | uidA | uidA + |
| 3 | Burkholderia cepacia | – | Negative (−) |
| 4 | Pneumoniae klebsiella | fimD | fimD + |
| 5 | Carbapenem-resistant enterobacteriaceae (CRE) | – | uidA + |
| 6 | Acinetobacter baumannii | – | Negative (−) |
| 7 | MRSA | mecA | nuc +, mecA+ |
| 8 | Enterococcus faecalis | – | Negative (−) |
| 9 | Enterococcus faecium | – | Negative (−) |
| 10 | Stenotrophomonas maltophilia | – | Negative (−) |
| 11 | Shigella | ipaH | ipaH+ |
| 12 | MRCNS | – | mecA+ |
| 13 | Staphylococcus epidermidis | – | Negative (−) |
| 14 | CRPA | – | Negative (−) |
| 15 | Candida albicans | IFA22 | IFA22+ |
| 16 | Negative control | – | Negative (−) |
+, indicates positive; −, indicates negative. MRSA, methicillin-resistant
Comparison of the coincidence rate of clinical samples and ordinary culture methods
From May 2019 to May 2021, a total of 278 clinical samples were collected at the Rocket Army Specialized Medical Center. Blood, urine, sputum, and drainage fluid were included in the sample types. In all, 24 clinical specimens of Staphylococcus aureus, 20 clinical specimens of Escherichia coli, 58 clinical specimens of Pneumoniae klebsiella, 34 clinical specimens of MRSA, 29 clinical specimens of Candida albicans, and 21 cases of simulated stool samples contaminated with Shigella were included. A total of 92 samples were negative for the target bacteria. Both negative and positive samples had been subjected to bacterial identification and drug susceptibility testing in accordance with the operating procedures. Meanwhile, they had been identified and confirmed by MALDI-TOF MS (Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry). In order to evaluate the performance of the detection system, negative and positive samples were tested in accordance with DNA extraction—LAMP and PCR reactions--Microfluidic Chips and Instruments—Operations on Chips. The results showed that the positive coincidence rates of Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, MRSA, and Candida albicans were 0.86, 0.9, 0.9, 0.95, 0.94, and 0.93, respectively. The negative coincidence rates were 0.92, 0.82, 0.71, 0.99, 0.91, and 0.74, respectively. Shigella showed excellent consistency with the microbial culture method, Staphylococcus aureus and MRSA showed excellent consistency, and Escherichia coli and Candida albicans showed general consistency, as shown in Table 7.
Table 7
| Staphylococcus, aureus (Sa) | Escherichia coli (E coli) | Pneumoniae klebsiella | Shigella | MRSA | Candida albicans | |
|---|---|---|---|---|---|---|
| Common positive (+) | 50 | 18 | 52 | 20 | 32 | 27 |
| Common negative (−) | 202 | 212 | 157 | 254 | 223 | 184 |
| Chip method (−), culture method (+) | 8 | 2 | 6 | 1 | 2 | 2 |
| Chip method (+), culture method (−) | 18 | 46 | 63 | 3 | 21 | 65 |
| Positive coincidence rate | 0.86 | 0.9 | 0.9 | 0.95 | 0.94 | 0.93 |
| Negative coincidence rate | 0.92 | 0.82 | 0.71 | 0.99 | 0.91 | 0.74 |
| Total coincidence rate | 0.91 | 0.83 | 0.75 | 0.99 | 0.92 | 0.76 |
| Random coincidence rate | 0.65 | 0.73 | 0.55 | 0.85 | 0.73 | 0.63 |
| Kappa value | 0.74 | 0.36 | 0.45 | 0.90 | 0.69 | 0.34 |
+, indicates positive; −, indicates negative. MRSA, methicillin-resistant
Discussion
With the use of broad-spectrum antibacterial drugs and immunosuppressive agents, the promotion of organ transplantation as well as the application of various indwelling catheters, people with low immune functions are more susceptible to multiple pathogen infections, which leads to prolonged hospitalization and increased hospitalization costs, thus adversely affecting the prognosis of the patient. Those patients in the intensive care unit are especially prone to infections from a variety of pathogens (1,2). With the widespread application of antibacterial drugs, the drug resistance of bacteria has continued increase. These pathogens need to be identified so that the targeted drugs can be used for the rehabilitation of patients, the alleviation of disease, and the rational use of antimicrobial drugs, especially when the bacteremia are severely endangered. It is particularly important to accurately detect the pathogenic infection as early as possible, so the correct antibacterial drugs can be applied (3,4). Currently, the detection of pathogenic bacteria in clinical practice mainly relies on conventional culture methods, such as the examination of body fluids, blood, and secretions (5,6); the cultivation and identification of pathogenic bacteria; and drug sensitivity tests. In recent years, research into the rapid detection of pathogens using molecular biology methods has made great progress (7-10). However, this detection requires expensive nucleic acid amplification equipment, with high requirements on laboratories and personnel, complicated procedures, and 3–5 hours of detection time. It is mainly suitable only in large medical institutions and professional laboratories. The gold standard for determining pathogenic bacteria, the traditional microbial culture method, involves a variety of instruments and reagents, also requires 3–5 days of detection time, and demands a high level of professional skills and experimental experience. Therefore, shortening the detection time and simplifying conditions have been strong focus of researchers in this field. LAMP is a new type of isothermal amplification method that was proposed by Notomi et al. (11). In this method, 4–6 specific primers are designed for 6 regions of the target gene, and the strand-displacement DNA polymerase is used at a Isothermal. Next, the DNA or RNA of the target genes are quickly amplified for detection via the observation of color change of the added indicator or by the intensity of fluorescent signal generated. The detection process only requires an incubator and no expensive and high-precision instruments, and the target gene can be amplified in large quantities and with high efficiency. This technique has low operational costs and a short detection time compared to classical polymerase chain reaction (PCR) techniques, and LAMP is highly selective for the target gene, which can reduce the influence of nontarget sequences, thus providing high specificity. The technology removes the traditional dependence on thermal cycle PCR instruments and can reduce the complexity of nucleic acid detection; therefore, the experimental results can be obtained within 1 hour. As microcasting processing technology has improved, researchers have intensified studies on microfluids (12) to analyze the research object using fluid in the micron-scale space. With the development of processing technology, the design of microchannels has been further optimized. Microfluidics can be divided into multiple reaction units for the detection of multiple pathogens, which can ensure the independence of each reaction and the accuracy of the results (13-19). The combined LAMP and microfluidics technology may be able to leverage the advantages of both technologies. The detection system requires only a small amount of samples and reagents, and the operating procedures are simple. There is no need for large and expensive instruments or professionally trained technicians. It can use imaging equipment such as cameras or the naked eye to observe the results. At present, the technology is mainly used in food safety, environmental pollution detection (20), criminal investigation (21), and inspection and quarantine (22,23). However, it is rarely used in large medical institutions, primary medical laboratories, and on-site disposal of infection events.
The rapid detection system combining microfluidic technology and LAMP technology established in this research can complete the nucleic acid extraction and detection of the original sample in a relatively short time. Through ROC curve analysis, the sensitivity of Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, MRSA, and Candida albicans at the best cutoff value was 86%, 85%, 88%, 95%, 97%, and 93%, respectively; the specificity was 93%, 89%, 74%, 100%, 90%, and 82%, respectively; and the area under the curve was 0.91, 0.91, 0.83, 0.97, 0.96, and 0.9, respectively; both system showed good diagnostic performance. Under the condition of determining the cutoff value, the detection limit and anti-interference ability of the detection system were analyzed. The detection limit of Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, MRSA, and Candida albicans was 6.95, 44.6, 3.89, 15.33, 16.45, and 463 pg/µL, respectively. In all, 15 kinds of bacteria were detected by the chip and were identified by the microbial mass spectrometer and microbial identification instrument, with any possible cross-reactions also being monitored. The results for 15 kinds of bacteria were positive in addition to the target bacteria, and the results of the other bacteria were negative, indicating that the detection of target bacteria using the microfluidic detection system did not cross-react with the detection of the other 15 kinds of strains, demonstrating the proposed method’s good anti-interference ability. In the analysis of the 278 clinical samples collected, including blood, urine, sputum, and drainage fluid, the positive coincidence rates of Staphylococcus aureus, Escherichia coli, Pneumoniae klebsiella, Shigella, and MRSA and Candida albicans were 0.86, 0.9, 0.9, 0.95, 0.94, and 0.93 respectively, and the relative negative coincidence rates were 0.92, 0.82, 0.71, 0.99, 0.91, and 0.74, respectively, while the total coincidence rates were 0.91, 0.83, 0.75, 0.99, 0.92, and 0.76, respectively. The consistency analysis with the bacterial culture method yielded Kappa values of 0.74, 0.36, 0.45, 0.90, 0.69 and 0.34, respectively. This indicated that the detection system for Shigella had excellent consistency with the microbial culture method, the detection of Staphylococcus aureus and MRSA had good consistency, and the detection of Escherichia coli Bacteria and Candida albicans was generally consistent. The performance of the detection system was assessed. Compared with that of the microbial culture method, the positive rate of the microfluidic detection chip was higher. This was likely related to the identification and drug sensitivity test of the dominant bacteria in the process of microbial culture, in which the dominant bacteria cannot be ruled out. There still remain other bacteria outside, and numerous similar studies have also confirmed this. Zhao (24) collected respiratory tract infection sputum specimens, applied the ordinary sputum culture, and used crystal core isothermal amplification technology for amplification analysis of common respiratory pathogens. The results showed that the positive rate of the crystal core respiratory pathogen detection was higher than that of the traditional sputum culture. The detection results of a Pneumoniae klebsiella, Streptomonas maltophilia, Legionella pneumophila. and Mycobacterium tuberculosis composite group were consistent. However, the detection rate of Streptococcus pneumoniae, Mycoplasma pneumoniae, Staphylococcus aureus, and Escherichia coli was significantly higher than that of traditional sputum culture, which helped to reduce missed detection and diagnose and treat the disease earlier. Li et al. (25) collected deep sputum samples from patients to detect pathogens using LAMP microfluidic chips and traditional isolation and culture methods. A total of 11 respiratory pathogens were detected, with a total positive rate of 58.27%. The positive rate of traditional isolation culture method was 35.8%, and 9 kinds of respiratory pathogens were detected.
Speaking generally, the LAMP microfluidic chip had a high detection rate, fast speed, high efficiency, and high sensitivity, which provided a reliable basis for rapid clinical diagnosis and precise treatment. One test can quickly detect a variety of pathogens, which is more advantageous than the application of more common sputum culture. Meanwhile, the LAMP microfluidic chip can directly analyze samples at the genetic level, which can save 16–18 hours of culture time. It can also analyze drug resistance genes synchronously, which helps clinicians and on-site infection personnel to make decisions.
Some limitations to our research should be addressed. First, there was diversity of sample types and a limited amount of samples. The accuracy of the results for different sample types can be further analyzed by increasing the sample size. In addition, in the compliance analysis of clinical samples for Shigella detection, the E. coli type in the simulated stool samples applied in the test was a normal flora, and the test was also positive. The accuracy in other sample types can be further explored in subsequent experiments.
In conclusion, LAMP technology and microfluidic chip technology can be combined to provide rapid detection of multiple pathogens. The system has good specificity, high sensitivity, strong anti-interference ability, and a relatively high coincidence rate in the detection of clinical samples. The equipment is simple and portable, with fast processing time and low cost, and is thus especially suitable for point-of-care testing (26). Identification of multiple infections of pathogens in medical institutions, especially in areas with suboptimal medical facilities, may not only reduce costs but may also provide the rapid diagnosis needed to make decisions quickly and confidently. This system may have considerable practical significance in the prevention and control of major diseases caused by pathogen infection and the treatment of public health disasters.
Acknowledgments
Funding: Project theme of Military Medical Science and Technology Youth Cultivation (16QNP030).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-2792
Data Sharing Statement: Available at https://dx.doi.org/10.21037/apm-21-2792
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-2792). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Y, Gong L, Gao A, et al. Analysis of the distribution characteristics and influencing factors of the departments of patients with multidrug-resistant bacteria infection. Journal of Clinical Emergency (China) 2019;29:3502-5.
- Liu S, Yang J, Liang X, et al. Clinical analysis of invasive fungal disease in emergency intensive care unit from 2013 to 2017. Journal of Clinical Emergency (China) 2017;18:511-4.
- Fernández J, Bert F, Nicolas-Chanoine MH. The challenges of multi-drug-resistance in hepatology. J Hepatol 2016;65:1043-54. [Crossref] [PubMed]
- Chen R, Guo G, Wu L, et al. Distribution and drug resistance analysis of pathogenic bacteria in hospital bloodstream infection. Guangdong Medicine (China) 2018;39:139-41.
- Dodémont M, De Mendonça R, Nonhoff C, et al. Performance of the Verigene Gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J Clin Microbiol 2014;52:3085-7. [Crossref] [PubMed]
- Fernández-Cruz A, Marín M, Kestler M, et al. The value of combining blood culture and SeptiFast data for predicting complicated bloodstream infections caused by Gram-positive bacteria or Candida species. J Clin Microbiol 2013;51:1130-6. [Crossref] [PubMed]
- Wen J, Gou H, Zhan Z, et al. A rapid novel visualized loop-mediated isothermal amplification method for Salmonella detection targeting at fimW gene. Poult Sci 2020;99:3637-42. [Crossref] [PubMed]
- Papadakis G, Murasova P, Hamiot A, et al. Micro-nano-bio acoustic system for the detection of foodborne pathogens in real samples. Biosens Bioelectron 2018;111:52-8. [Crossref] [PubMed]
- Park BH, Oh SJ, Jung JH, et al. An integrated rotary microfluidic system with DNA extraction, loop-mediated isothermal amplification, and lateral flow strip based detection for point-of-care pathogen diagnostics. Biosens Bioelectron 2017;91:334-40. [Crossref] [PubMed]
- Heymans R, Vila A, van Heerwaarden CAM, et al. Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS One 2018;13:e0206316 [Crossref] [PubMed]
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000;28:E63 [Crossref] [PubMed]
- Reyes DR, Iossifidis D, Auroux PA, et al. Micro total analysis systems. 1. Introduction, theory, and technology. Anal Chem 2002;74:2623-36. [Crossref] [PubMed]
- Liu N, Zou D, Dong D, et al. Development of a multiplex loop-mediated isothermal amplification method for the simultaneous detection of Salmonella spp. and Vibrio parahaemolyticus. Sci Rep 2017;7:45601. [Crossref] [PubMed]
- Shang Y, Sun J, Ye Y, et al. Loop-mediated isothermal amplification-based microfluidic chip for pathogen detection. Crit Rev Food Sci Nutr 2020;60:201-24. [Crossref] [PubMed]
- Zhou QJ, Lu JF, Su XR, et al. Simultaneous detection of multiple bacterial and viral aquatic pathogens using a fluorogenic loop-mediated isothermal amplification-based dual-sample microfluidic chip. J Fish Dis 2021;44:401-13. [Crossref] [PubMed]
- Wan L, Gao J, Chen T, et al. LampPort: a handheld digital microfluidic device for loop-mediated isothermal amplification (LAMP). Biomed Microdevices 2019;21:9. [Crossref] [PubMed]
- Mao R, Ge G, Wang Z, et al. A multiplex microfluidic loop-mediated isothermal amplification array for detection of malaria-related parasites and vectors. Acta Trop 2018;178:86-92. [Crossref] [PubMed]
- Yuan H, Chao Y, Shum HC. Droplet and Microchamber-Based Digital Loop-Mediated Isothermal Amplification (dLAMP). Small 2020;16:e1904469 [Crossref] [PubMed]
- Azizi M, Zaferani M, Cheong SH, et al. Pathogenic Bacteria Detection Using RNA-Based Loop-Mediated Isothermal-Amplification-Assisted Nucleic Acid Amplification via Droplet Microfluidics. ACS Sens 2019;4:841-8. [Crossref] [PubMed]
- Wu J, Huang L, Wang Y, et al. Application of microfluidic chip electrophoresis in food safety and environmental pollution detection. Chinese Journal of Analysis Testing 2015;34:283-8.
- Ashraf W, Unger H, Haris S, et al. Genetic detection of peste des petits ruminants virus under field conditions: a step forward towards disease eradication. BMC Vet Res 2017;13:34. [Crossref] [PubMed]
- Jv H, Dai J, Xie Y, et al. LAMP microfluidic chip for rapid detection of three food-borne pathogens. Chinese Journal of Preventive Medicine (China) 2018;36:309-313.
- Meng X, Zhang G, Sun B, et al. Rapid Detection of mecA and femA Genes by Loop-Mediated Isothermal Amplification in a Microfluidic System for Discrimination of Different Staphylococcal Species and Prediction of Methicillin Resistance. Front Microbiol 2020;11:1487. [Crossref] [PubMed]
- Zhao L. Crystal core respiratory pathogen nucleic acid detection technology and sputum culture in clinical application. Journal of Practical Laboratory Medicine (China) 2019;11:162-4.
- Li Y, Gao M, Sai Y, et al. The value of thirteen combined detection of respiratory pathogen nucleic acid Isothermal amplification chip in the diagnosis of lower respiratory tract infection. Chinese Journal of Lung Diseases 2019;12:171-4. (Electronic Edition) (China).
- Hua W, Sheng L, Wang Q. Progress in the application of POCT in the diagnosis of infectious diseases. Chinese Journal of Laboratory Medicine (China) 2019;333-7.