
Global trends and hotspots in research on organoids between 2011 and 2020: a bibliometric analysis
Introduction
Organoids are defined as 3D structures of stem cells that contain different organ-specific cell types. Owing to the wide range of tissue types, the long-term expansion capacity, and the physiological 3D architecture, organoids are a powerful new technology for many biological and clinical applications. Organoids have been used as models of infectious (1,2) and genetic (3,4) diseases, as well as cancer (5,6). Brain organoid technology has been used in developing drugs to prevent or treat Zika infection (7); tumor organoids provide high-throughput chemotherapeutic drug screening platforms (8) and are also conducive to predicting the treatment response for metastatic gastrointestinal cancers (9). The gene correction of cystic fibrosis transmembrane conductance regulator (CFTR) mutation in patients derived organoids using CRISPR/Cas9 gene editing could repair the CFTR function and treat cystic fibrosis (10). Organoid technology also holds great promise for personalized medicine, as organoids can be used for rapid, ex vivo testing of drug responses in diseased tissues or organs of individual patients. In addition, the biological banks can be served as resources for studying cancer biology and drug development. These living organoid biobanks have been established for multiple organs and tumor types, including histologically and genetically characterized tumors and matching normal organoids (11,12). Furthermore, organoids could potentially be investigated for the development of alternative therapy strategies for organ transplantation, such as intestinal and liver organoid transplantation (13,14). In recent years, organoid publications have increased, suggesting that many researchers utilize organoid technology in various fields. However, this rapid expansion of the literature can make it difficult to provide an overall perspective on the field’s state and the emerging trends using traditional review methods.
At present, most of the related studies remain scattered, and prove unfavorable to clinical transformation and for serving improved clinical treatment. This requires us to summarize the progress in the existing fields, in order to meet clinical needs. For example, the protocol for culture varies in different systems. An objective summary and analysis of the publications can lead to a standardized organoid culture protocol. Further, summarizing organoid-related publications can contribute to finding out the limitations and the cause of the limitations; for instance, current organoids are not suitable for the screening of immunotherapeutic agents owing to the lack of surrounding stromal cells (15). In addition, the summary and analysis of the publications can help identify possible future solutions to overcome current clinical shortcomings; for example, the combination of organoid and engineering technologies may be the future approach to exploit their potential in clinical applications (16).
The bibliometric analysis is based on statistics and visualization techniques. It is a valuable method to evaluate quantitatively the influence of research papers in a certain period, considering countries, institutions, journals, and research collaborations for any specific topic while measuring developmental research trends and depicting knowledge structures related to a research discipline over time (17,18). Using bibliometrics, investigators can quickly identify the key articles worthy of intensive, careful review among a large number of documents, excavate the frontiers of disciplines, and pinpoint research hotspots. This technique also offers an objective method to determine the influence and importance of disciplines and topics, study the distribution of subject or topic information sources, identify core journals, examine the needs and characteristics of information users, evaluate talents, and visualize data. Bibliometric analyses contain many science mapping tools, which adopt timely, repeatable, and flexible methods to produce a complex interactive visual structure for statistical analyses (19). Traditional reviews cannot accomplish these aims because of the unmanageable manual workload. Moreover, the results of bibliometric analyses are determined by specific indicators rather than the intervention of experts and prior working knowledge or experience in the field. Bibliometric analyses can also be verified by any analyst and repeated. Therefore, bibliometric analyses enable the objective metrological study of organoids. The bibliometrix software package, VOSviewer, CiteSpace BICOMB, and gCLUTO are five frequently used bibliometric tools that allow scientists to evaluate the current state of knowledge in a subject area and identify related hotspots (20,21). Many hotspots on organoids have appeared recently, such as developing novel drugs for tumor patients and predicting patients’ responses to individualized treatments using organoids. To the best of our knowledge, this is the first scientometric study that describes organoid research’s current and future directions.
We conducted a quantitative analysis of the literature, considering author contributions, country/region/institution/author co-authorship, journal/reference co-citation analysis, document citation analysis, co-occurrence of keywords, thematic evolution, keywords and references with strong citation burstness, co-word biclustering analysis, and landform map. This study aimed to provide an overview of the field, elucidate the trends in different aspects of organoid research, and point out the limitations of existing research while proposing possible solutions.
Methods
Data source and search strategy
On Oct 4, 2021, we used the Web of Science (WoS) core collection Science Citation Index expanded (SCI-EXPANDED) database (Thomson Reuters, New York, NY, USA) to conduct a literature search and identify all types of documents related to organoids published in the past decade (from 2011 to 2020), with no language restrictions. The search strategy syntax contained “organoid” OR “organoids”, searched in topic term. Three authors screened the retrieved literature independently and determined eligibility. Finally, no related documents were excluded. The search identified 3,168 publications (articles and reviews). The full data, including author, title, abstract, keywords, source, language, citation, etc., was downloaded as a BibTeX and txt file from the WoS database. The publications were then processed with the bibliometric tools described below.
Statistical analysis
General analysis and thematic evolution analysis
The general basic characteristics of the retrieved publications, including the country/region, the year of publication, and the authors, were converted and analyzed automatically using the bibliometrix software package in R 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria) and Microsoft Excel (version Microsoft 365). Journal impact factors were obtained from the 2020 Journal Citation Reports (Clarivate Analytics, Philadelphia, PA, United States). To evaluate the quality of the documents, we used the number of publications and citations in the related fields, the h-index value, and the m-index value. The h-index can be used to evaluate the performance by individual citation analysis of journals/references/citations of documents and co-occurrence of keywords or by groups of scientists working in university departments or research institutes. The m-index facilitates comparing scholars with different academic career lengths (22). In addition, the Bibliometrix software package was used to create a thematic evolution of 10 years, divided into three periods (2011–2014, 2014–2017, and 2017–2020). This enabled the display of changing time-trends of keywords, for those three periods. Furthermore, GraphPad Prism 9.0 was used to conduct nonlinear regression (curve fit) for the trend of the number of publications in this field at each point.
Network analysis
VOSviewer (Version 1.6.16, Leiden University, the Netherlands) was used to analyze organoid research collaboration networks and co-citation networks, as well as to identify research hotspots and future trends. We generated networks connecting productive countries/regions/institutions (if one publication was attributed to authors from more than one country/region, the publication was assigned equally to all participating countries/regions), authors, and co-authors using co-authorship, co-occurrence, citations, and co-citation analysis. Specifically, overlay mapping was conducted to show the time scale of themes in the organoid field. In the visual maps, different colors represented different clusters, and connecting lines indicated collaboration or co-citations. The numbers of documents, citations, and keyword occurrences were represented by circle size, whereas the strength of the links was represented by the thickness of the connecting lines. In the co-occurrence analysis, keywords that occurred more than five times were presented in three visualizations (network, overlay, density visualization) in the co-occurrence networks.
Burstness analysis
CiteSpace (5.8.R5) (http://cluster.cis.drexel.edu/~cchen/citespace/) is a bibliometric tool that utilizes Java to visualize and analyze trends and patterns in scientific documents. We used it to search for keywords and references with strong citation burstness and identify the latest research focus and future directions in organoid research. For burstness analysis visualizations, the parameters of CiteSpace were set as follows: time slicing [2010–2020], years per slice (1 year), and visualization (time zone view). A more detailed description of the software, utilization skills, and options can be found in the CiteSpace manual. The keywords and references surging in the corresponding period suggested the changing trend of possible research focuses.
Co-word biclustering analysis and landform map
The data were imported into BICOMB (23) to construct a keyword-keyword binary matrix (21). The rows and columns represented highly frequent keywords (n>20). Additionally, the gCLUTO software 1.0 was used to perform the clustering analysis; keywords in each cluster may represent a topic. Landform maps and heatmaps were also generated based on the results of the clustering analysis.
Results
Trends in global publications
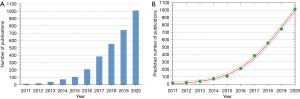
A total of 3,168 articles related to organoids were retrieved from WoS. Global literature in this field exhibited a strongly increasing trend, going from 13 articles (0.41%) published in 2011 to 1,010 articles (31.9%) published in 2020 (Figure 1A). Using logistic regression analysis, we constructed a curve of publications per year, which revealed that this field is currently at the stage of stable growth with respect to global publications output (Figure 1B).
Distribution of countries/regions and institutions
To visualize the publication contributions of different countries/regions to organoid-related research, we used R to prepare a world map of publication productivity (Figure 2A). A total of 39 countries and regions contributed to publications in this field. The plurality of documents was published by groups based in the United States (1,063, 33.55% of all articles), followed by China (223, 7.04%), the Netherlands (163, 5.15%), Japan (145, 4.58%), and Germany (118, 3.72%) (Figure 2B). The highest number of citations emanated from the United States (22,571 citations), followed by the Netherlands (9,995 citations), the United Kingdom (4,659 citations), Japan (4,549 citations), Austria (4,529 citations), and Germany (2,761 citations) (Figure 2C).

Next, we conducted a co-authorship analysis of 19 countries/regions that had produced more than 20 publications in this field (Figure 3A). The five countries/regions with the highest total link strength among authors were the United States (total link strength =472 times), the Netherlands (227 times), England (200 times), Germany (198 times), and China (157 times).
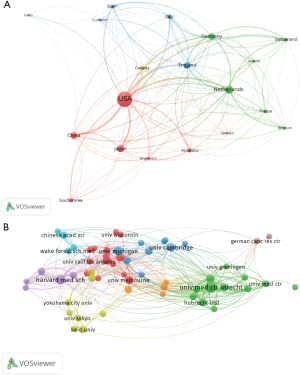
At the time of our analysis, a total of 2,696 organizations were involved in this field. The University of Michigan had the highest number of publications (290 records, or 10.76% of all articles), followed by the University Medical Center Utrecht (268, 10.76%), the University of Cambridge (196, 7.27%), the University of Pennsylvania (177, 6.57%), and Johns Hopkins University (172, 6.40%).
Finally, we analyzed the co-authorship relationships of 59 institutions with more than 20 publications using viewOS tools. Only one document was excluded (because it was not connected to the others), which suggests that a network of collaboration exists between these 59 institutions (Figure 3B). The University Medical Center Utrecht had the greatest total link strength (total link strength =167), followed by the Princess Máxima Center for Pediatric Oncology (total link strength =99), Harvard Medical School (total link strength =96), Universiteit Utrecht (total link strength =87), and the Royal Netherlands Academy of Arts and Sciences (total link strength =79).
Analysis of journals
In all, 3,168 publications were obtained from 696 sources. The top 10 most popular journals related to organoids are listed in Table 1. Cancer Research (175 records, 5.52% of all articles) had the most publications, followed by Gastroenterology (114, 3.60%), Investigative Ophthalmology & Visual Science (82, 11.79%), Scientific Reports (69, 9.91%), and Cell Stem Cell (66, 9.48%).
Table 1
| Rank | Popular journals | Record (n) | 2020 impact factors | 2020 JCR partition | Cited journals | Citations (n) | 2020 impact factors | 2020 JCR partition |
|---|---|---|---|---|---|---|---|---|
| 1 | Cancer Research | 175 | 9.727 | q1 | Nature | 7,122 | 42.778 | q1 |
| 2 | Gastroenterology | 114 | 17.373 | q1 | Cell | 4,777 | 38.637 | q1 |
| 3 | Investigative Ophthalmology & Visual Science | 82 | 3.528 | q1 | Cell Stem Cell | 3,864 | 20.860 | q1 |
| 4 | Scientific Reports | 69 | 3.998 | q1 | P Natl Acad Sci USA | 3,227 | 9.412 | q1 |
| 5 | Cell Stem Cell | 66 | 20.860 | q1 | Science | 2,994 | 41.845 | q1 |
| 6 | Pediatric Pulmonology | 60 | 2.534 | q1 | Development | 2,151 | 5.611 | q2 |
| 7 | FASEB Journal | 46 | 4.966 | q1 | Nat Med | 2,117 | 36.13 | q1 |
| 8 | Jove-Journal of Visualized Experiments | 46 | 1.163 | q3 | PLoS One | 1,742 | 2.74 | q2 |
| 9 | Cancer Science | 44 | 4.966 | q1 | Nat Biotechnol | 1,734 | 36.558 | q1 |
| 10 | Tissue Engineering Part A | 42 | 3.496 | q2 | Nat Commun | 1,657 | 12.121 | q1 |
JCR, Journal Citation Reports; P Natl Acad Sci USA, Proceedings of the National Academy of Sciences of The United States of America; Nat Med, Nature Medicine; Nat Biotechnol, Nature Biotechnology; Nat Commun, Nature Communications.
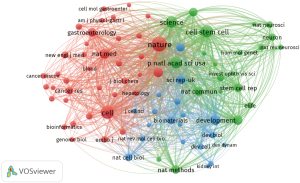
We analyzed a total of 76 journals for publications that were co-cited in more than 200 other publications (Figure 4). Table 1 displays the top 10 journals for citations in this field. Nature was cited the most (7,122 citations), followed by Cell (4,777 citations), Cell Stem Cell (3,864 citations), Proceedings of the National Academy of Sciences of the United States of America (PNAS) (3,277 citations), and Science (2,994 citations).
Analysis of authors
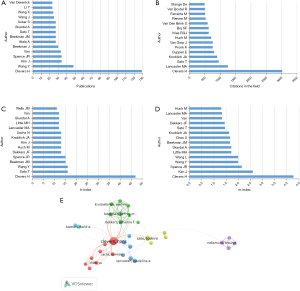
In terms of the number of publications, Hans Clevers is the top author, with 129 related articles (4.07% of all articles), followed by Yaqing Wang (48, 1.52%), Jihoon Kim (34, 1.07%), Jason R. Spence (34, 1.07%), and Marc van de Wetering (33, 1.04%) (Figure 5A). In terms of the number of citations in this field, Clevers was again the first (3,021 citations), followed by Madeline A. Lancaster (1,249 citations), Toshiro Sato (952 citations), Juergen A. Knoblich (898 citations), and Edwin Cuppen (801 citations) (Figure 5B). Clevers had the highest h-index [47], followed by those from Sato [16], Wang [16], Jeffrey M. Beekman [15], and Spence [15] (Figure 5C). The m-index of publications from Clevers (4.273) was also ranked first, followed by those from Kim (2.600), Spence (2.143), Yaqing Wang (2.000), and Li Wang (2.000) (Figure 5D). We analyzed a total of 74 authors who were co-authors on more than 10 publications (Figure 5E). Excluding 42 authors, who were not connected, 32 demonstrated collaborative links. The five authors with the highest total link strength were Clevers (total link strength =115), Beekman (total link strength =81), Cornelis K. van der Ent (total link strength =75), Johanna F. Dekkers (total link strength =71), and Karin M. de Winter-de Groot (total link strength =63).
Citation and co-citation analyses
Next, the citation analysis revealed that 132 documents had more than 100 citations (Figure 6A). Table 2 lists the ten documents with the highest number of citations. There were 1,872 citations for “Cerebral organoids model human brain development and microcephaly” (24), followed by “Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium” (25), which was cited 1,463 times. The third-ranked article for the number of citations was “Organogenesis in a dish: Modeling development and disease using organoid technologies” (26), which had 1,001 citations.

Table 2
| Rank | Title | Authors | Source | Publication, year | Citations (n) |
|---|---|---|---|---|---|
| 1 | Cerebral organoids model human brain development and microcephaly | Lancaster et al. (24) | Nature | 2013 | 1,872 |
| 2 | Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium | Sato et al. (25) | Gastroenterology | 2011 | 1,463 |
| 3 | Organogenesis in a dish: Modeling development and disease using organoid technologies | Lancaster, Knoblich (26) | Science | 2014 | 1,001 |
| 4 | Modeling development and disease with organoids | Clevers (27) | Cell | 2016 | 867 |
| 5 | Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure | Qian et al. (28) | Cell | 2016 | 865 |
| 6 | Prospective derivation of a living organoid biobank of colorectal cancer patients | van de Wetering et al. (29) | Cell | 2015 | 805 |
| 7 | Organoid models of human and mouse ductal pancreatic cancer | Boj et al. (30) | Cell | 2015 | 793 |
| 8 | Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients | Schwank et al. (31) | Cell Stem Cell | 2013 | 729 |
| 9 | Organoid cultures derived from patients with advanced prostate cancer | Gao et al. (11) | Nature | 2014 | 663 |
| 10 | Zika virus impairs growth in human neurospheres and brain organoids | Garcez et al. (32) | Nature Cell Biology | 2016 | 604 |
CFTR, cystic fibrosis transmembrane conductance regulator; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9.
We analyzed 52 references co-cited in more than 100 citations (Figure 6B). Table 3 lists the top 10 references with the highest number of citations. The five references with the largest number of citations were by Sato et al. (33) (2009, Science; 574 citations), Lancaster et al. (24) (2013, Nature; 453 citations), Sato et al. (25) (2011, Gastroenterology; 438 citations), van de Wetering et al. (29) (2015, Cell; 319 citations), and Lancaster, Knoblich (26) (2014, Science; 313 citations).
Table 3
| Rank | Title | Authors | Source | Publication, year | Citations (n) |
|---|---|---|---|---|---|
| 1 | Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche | Sato et al. (33) | Nature | 2009 | 574 |
| 2 | Cerebral organoids model human brain development and microcephaly | Lancaster et al. (24) | Nature | 2013 | 453 |
| 3 | Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium | Sato et al. (25) | Gastroenterology | 2011 | 438 |
| 4 | Prospective derivation of a living organoid biobank of colorectal cancer patients | van de Wetering et al. (29) | Cell | 2015 | 319 |
| 5 | Organogenesis in a dish: modeling development and disease using organoid technologies | Lancaster, Knoblich (26) | Science | 2014 | 313 |
| 6 | Modeling Development and Disease with Organoids | Clevers (27) | Cell | 2016 | 293 |
| 7 | Organoid models of human and mouse ductal pancreatic cancer | Boj et al. (30) | Cell | 2015 | 268 |
| 8 | Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro | Spence et al. (34) | Nature | 2011 | 249 |
| 9 | Organoid cultures derived from patients with advanced prostate cancer | Gao et al. (11) | Cell | 2014 | 243 |
| 10 | Vascularized and functional human liver from an iPSC-derived organ bud transplant | Takebe et al. (35) | Nature | 2013 | 216 |
Lgr5, leucine rich repeat containing g protein-coupled receptor 5; iPSC, induced pluripotent stem cell.
Co-occurrence analysis and thematic evolution analysis
We analyzed 303 keywords that were identified as occurring more than 10 times (Figure 7A). The colors in the overlay visualizations shown in Figure 7B,7C indicate the average publication year of the identified keywords. Most co-occurring keywords analyzed from 2011 to 2020 that were cited at least 20 times were published after 2018.4, as indicated by the greener and yellower colors on the network map. The density visualization showed the same keywords mapped by frequency of appearance and revealed that the top five co-occurring keywords were “in vitro”, “pluripotent stem cells”, “differentiation”, “stem cells”, and “organoids” (Figure 7D). To better understand the evolution of the research focus, we performed thematic evolution analysis, as shown in Figure 7E. This analysis was performed by dividing the decade into three periods and visualizing the important keywords for each period and their evolution. Keywords such as “tissue engineering”, “lgr5”, “mammary gland”, “Cohn’s disease”, “3D culture”, and “inflammatory bowel disease” emerged in organoid research before 2014. New keywords, such as “developmental biology”, “patient-derived organoid”, “Zika virus”, “glioblastoma”, and “cystic fibrosis” emerged in 2018–2020, thus creating some new focus vocabulary. The keywords “lgr5” and “3D culture” in the past suggested that the theme at that time was how to build an organoid model. However, the practical and extensive applications of complex organoids are hotspot at present. For example, developmental biology and disease modeling from patient-derived organoids to treat diseases, such as Zika virus (ZIKV), glioblastoma, and cystic fibrosis, have become new hot topics in recent years.
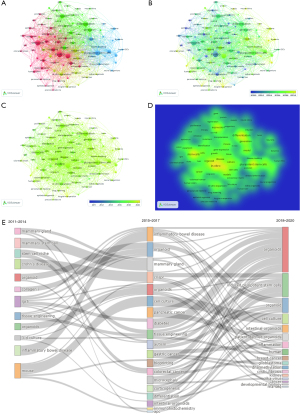
Burstness analysis of keywords and references
The burst detection allows researchers to identify publications and keywords that have received particular attention from related scientific communities during a certain period. Using CiteSpace to conduct the burstness analysis of keywords, we identified 120 keywords with strong citation burstness, indicating a trend of popular keywords from 2011–2020 (Figure 8A). The red rectangle on the right side of Figure 8A indicates a surge in the number of citations of the keyword at this stage. The most recent citation bursts of keywords, such as developmental biology, microenvironment, and xenograft, represent the most recent research focus areas in the field. In addition, our burst detection analysis also identified 90 articles with strong citation burstness (Figure 8B). The most recent citation bursts of publications were from eight articles whose bursts ended in 2020; among them, publications with relatively strong burstness were published in Nature Protocols, Nature Communications, Nature, Development, Gut, Mucosal Immunology, and Assay and Drug Development Technologies.
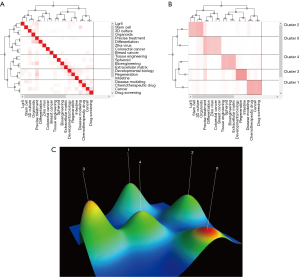
Co-word biclustering analysis and landform map
BICOMB and gCLUTO were performed to identify the correlation degree of each keyword and classify them into five clusters. The connections within clusters were also displayed by dendrograms on the axes. The darker the red, the higher the degree of correlation. Interestingly, each cluster could be grouped into a theme. Cluster 0 represented precise treatment; cluster 1 represented disease modeling and drug screening; cluster 2 represented methods to build organoid systems; cluster 3 represented developmental biology; cluster 4 represented tissue engineering and bioengineering (Figure 9A,9B). A 3D landform map was generated to reveal the inter-cluster standard deviation (Figure 9C). The curve of each mountain peak was a Gaussian curve, which approximately reflected the distribution of the data in the associated cluster. Position and height reflected the inter-cluster similarity, whereas the volume reflected the number of terms within the cluster. The most meaningful information was the color of the peak, which revealed the inter-cluster standard deviation; red indicated low deviation, while blue denoted high variance.
Discussion
Using a group of research-based articles on organoids retrieved from the WoS database, published between 2011 and 2020, we performed bibliometric analyses combined with network visualizations to obtain a comprehensive view of the current research trends concerning organoids and provide a reference for researchers in this field. To this end, we analyzed the contributions of countries or regions, organizations, journals, and authors to this emerging field and predicted hot topics that warranted further research. Since 2011, the annual number of publications related to organoid research has increased sharply, suggesting that the enormous potential inherent in organoid technology may have inspired researchers to make increasing use of this exciting technique. Indeed, we believe that organoids hold the potential to revolutionize disease research profoundly.
Our analyses revealed that the USA had the highest number of publications and citations and ranked first in the national co-author analysis; hence, the USA is the clear leader in this field, with a significant impact and the greatest collaborative activity with other countries/regions in organoid research. The number of studies on organoids conducted in China (223, 7.04%), the Netherlands (163, 5.15%), Japan (145, 4.58%), and Germany (118, 3.72%) increased significantly over the years, accounting for 20.49% of all included studies; thus, these five countries are particularly influential in the field of organoid research. Among the top 15 most influential countries/regions in organoid research, eight were European, which suggests that European research capacity is relatively strong. China ranks fourth, sixth, and fifth in the total number of papers published, the total number of citations, and cooperation with other countries/regions, respectively. The University of Michigan is the most productive in terms of organoid research, and the University Medical Center Utrecht ranks first in the co-authorship analysis, which suggests that it cooperates closely with other organizations.
Cancer Research has published the highest number of organoid-related articles among all journals in the field of organoid research, while Nature is the most co-cited journal in the field, with 7,122 citations. In addition, the top journals Cell and Science were ranked second and fifth in the co-cited list, respectively. Cell Stem Cell was the only journal in the top 10 for both productiveness and co-citation.
Hans Clevers of the University Medical Center Utrecht in the Netherlands is an outstanding scientist in the field of organoid research. He ranks first in the number of articles published, and his articles have been cited over 3,000 times, which is more than twice as many citations as the next author on the list; hence, he ranks first among authors in terms of h-index, m-index, and co-authorship analysis results. Clevers’ h-index and m-index results were 47 and 4.273, respectively, which indicates that this author has made a major contribution to the literature and that he is a pioneer and key author in organoid research.
Influential studies
A publication’s number of citations often reflects its academic influence. We analyzed the 10 most frequently cited articles in the field of organoid research, demonstrating significant advances in organoid research, using the VOSviewer tool. The study with the highest number of citations (n=1,872) was published in Nature in 2013 (24). It demonstrated a novel approach to studying human neurodevelopmental processes by using human pluripotent stem cells to construct and culture cerebral organoids in vitro. This method recapitulates the fundamental mechanisms of mammalian neurodevelopment and displays the characteristics of human brain development. In addition, these authors modeled microcephaly using these cerebral organoids, which suggested that organoids could be used to provide novel insights into the pathogenesis of neurological diseases. In 2011, Sato et al. (25) published an article in Gastroenterology, which ranked second (n=1,463) in terms of the number of citations. The authors reported that they used the replicative potential of adult stem cells to construct organoids that could be used to study infected, inflammatory, or neoplastic tissues from the human gastrointestinal tract. A review by Lancaster and Knoblich, published in Science in 2014, was cited 1,001 times, ranking third in the top 10 cited articles (26). This article summarized the rapidly growing field of organoids and outlined the future potential of organoid technology in biomedical research. The authors hypothesized that disease models could be the main focus of organoid research in the future and that organoids could also be used to test the efficacy and toxicity of drug compounds and even in whole-organ replacement in the clinic. Clevers (27) published the fourth-most cited review (n=867) in Cell in 2016, wherein he described the state of the exponentially developing field of induced pluripotent stem cell (iPSC)- and ASC-based organoids. Clevers also pointed out that current modes of preparation of organoids had some limitations, such as their lack of blood vessels and immune cells, which results in only the partial recapitulation of disease processes. The fifth-most cited article (n=865) was published by Qian et al. (28). They invented a miniaturized spinning bioreactor to culture brain-region-specific organoids based on human iPSCs. These authors then used organoids to model the exposure of the forebrain to the ZIKV, demonstrating that both African and Asian ZIKV strains preferentially and productively infect neural progenitors in organoids. Their organoids and bioreactor provided an accessible and versatile platform for compound testing of the ZIKV. Next, van de Wetering et al. (29) published the sixth-most cited study in Cell in 2015, with 805 co-citations; it reported the construction of tumor organoid cultures from 20 consecutive colorectal carcinoma (CRC) patients. The authors showed that these tumor organoids closely recapitulated several properties of CRC, such as somatic copy number and mutation spectra, and the organoids were amenable to high-throughput drug screening, enabling the design of personalized therapy. In 2015, Boj et al. (30) published the seventh-most cited article, also in Cell (n=793). They reported the establishment of pancreatic organoids as a tractable and transplantable system with which to investigate the molecular and cellular characteristics of mouse and human tumor progression. In particular, the authors successfully generated a normal ductal architecture after orthotopic transplantation. They also found that nucleoporins are broadly upregulated in neoplastic murine organoids, which indicates that they could be associated with the initiation of cancer development. The eighth-most cited article was published in Cell Stem Cell by Spence et al. (34). This article illustrated that a series of growth factors, such as WNT3A and FGF4, can be manipulated to direct the differentiation of human iPSCs into intestinal tissue and mimic embryonic intestinal development in vitro. The ninth-most cited article (n=663) was published in PNAS in 2005 by Gao et al. (11), who reported the successful construction of a long-term culture of prostate cancer organoids from biopsy specimens and circulating tumor cells. They also generated models of key prostate-specific genetic alterations, including ETS-translocations, SPOP mutations, FOXA1 mutations, and CHD1 loss. The tenth-most cited article (n=604) was published in Nature Cell Biology and was written by Garcez et al. (32). In this article, the authors examined the influence of ZIKV infection on human neural stem cells forming brain organoids, which suggests that ZIKV damages neurogenesis during human brain development.
Knowledge base
The number of co-cited articles represents how frequently at least two publications are cited together by other publications and thus can be considered a knowledge base for a specific field or subject (36). Our study selected the top 10 co-cited references to define the knowledge base related to organoid research. Among them, eight articles were also found on the list of the top 10 cited references as well as the list of the top 10 co-cited references, which indicated that these publications were not only part of the knowledge base but were also considered influential studies. The other two publications were “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche” and “Vascularized and functional human liver from an iPSC-derived organ bud transplant”. The former is the most co-cited article (n=574) and was published by Sato et al. (33). In it, the authors illustrated that a single Lgr5+ intestinal stem cell could operate independently of positional cues from its environment and generate a sustained, expanding, self-organizing epithelial structure similar to a normal gut. The latter paper is the tenth-most co-cited article, published by Takebe et al. (35), who used human iPSCs to generate liver buds in vitro (iPSC-LBs), which can eventually differentiate into a vascularized and functional human liver after transplantation.
Hot topics, limitations of existing studies, and possible future solutions
According to our analysis, the main current hot topics in organoid research concern drug screening (29,37-39), disease modeling (used to identify human-specific disease mechanisms) (24,26,40-42), personalized medicine/ precision medicine (43,44), tissue engineering (regenerative medicine) (45-47), developmental biology (48-51). These topics are likely to be the key research directions in the future. However, there are still some limitations in the current organoid research, which concern the lack of reproducibility, the limited level of maturity and function, and the absence of appropriate functional readouts. The lack of reproducibility is the most severe problem. This is because the formation of organoids mainly depends on a self-organization principle with minimal control over the external inputs supplied to the system. In addition, the uncontrolled nature of these processes leads to high heterogeneity in the current organoid systems among different laboratories (52). Thus, achieving robust and reproducible organoid cultures is a priority, as only then will it be possible to utilize them in basic and clinical studies. Applying bioengineering strategies to control organoids’ micro-environments by integrating chemical (such as the chemical modification of hydrogels) and physical instructions (such as geometry and mechanical properties) could be essential to control organoid self-organization and differentiation. Thus, instead of culturing organoids without any constraints, researchers should adopt some biomimetic hydrogels (53) and well-defined engineering technologies, such as micropatterning techniques (54), extracellular matrix (ECM) dynamics (55), and microfluidic technology (56), to ensure the homogeneity of organoids in a highly controlled manner-both spatiotemporally and in terms of dosage. Further studies are required to realize the potential of these bioengineering strategies in the organoid field. Furthermore, standardized protocols with a tendency toward more complex systems should be developed to achieve superior homologous organoids.
Another significant limitation is the reduced level of maturity and function. None of the current organoid systems can mimic the full functional repertoire of the organs. The main reasons are the limited lifespan of organoids, the loss of a mesenchymal compartment, vascularization, and/or microbiome. For example, the limited lifespan leads to dead cells accumulating in the hollow lumen in cystic epithelial organoids (24). Brain organoids generally fail to mature beyond a fetal phenotype (57). The potential solution to this shortcoming could be the promotion of vascularization, tissue-tissue interactions, and mimicking physiological environments. Vascularization in organoids enables the convective transport of nutrients/gases and waste removal, increasing the organoid lifespan and the size and complexity compared to static culture systems (58), while it is also difficult to achieve stable and mature microvascular networks within organoids. Engineering tissue-tissue interactions could be another solution to increase the level of organoid maturity since the morphology of organs is mediated by signals from adjacent tissues. Some examples are the generation of intestinal tissues with a functional enteric nervous system (59) and the recapitulation of human hepato-biliary-pancreatic tissue-tissue boundaries (60), although these approaches need optimization to achieve organoid to organoid integration with the ability to recapitulate the development in vivo. Some micro-technology, such as the organ-on-a-chip, can mimic physiological-like environments and theoretically capture key aspects of (human) organ physiology. In the future, coupling organ-on-a-chip systems may increase our understanding of complex inter-organ behaviors in health versus disease scenarios.
The absence of appropriate functional readouts is also a crucial limitation. Currently, organoid research mainly depends on phenotypic readouts, including aspect, shape, and number of organoids, which provide only limited information about the functionality of the organoids. Thus, continuous, accurate, and versatile functional readouts, such as image-based analyses (61), in situ electrochemical probing (62), and high-throughput setups (63), will enable the detection of subtle cellular responses and achieve a precise characterization of organoids in the future. These solutions are likely to become hot topics in the future, as they promote the further application of organoids for drug screening, disease modeling, and personalized medicine/ precision medicine.
Strengths and limitations
To the best of our knowledge, this is the first investigation to employ measurement analysis of organoid research trends. Publications in this field were collected and fully investigated by quantitative and qualitative analysis, as was research quality across different authors, using the R bibliometrix package. Two well-known scientometric software tools (VOSviewer and CiteSpace) were used to construct and visualize the bibliometric networks through co-authorship, co-citation, and co-occurrence analyses, and the top 90 articles with the strongest citation bursts were identified. However, our study has some limitations. First, our searches were only conducted in the WoS database, and the accuracy of our analyses could be improved by accessing other databases as well. Second, the results obtained from the bibliometric analysis may not be comprehensive; it is possible that some information considered important in this field may have been published in articles other than those in our analyses. Finally, it is possible that we have not paid sufficient attention to some important recent publications and have not discussed them in sufficient detail.
Conclusions
Our results indicate that the United States has made the largest contribution to organoid research, while Hans Clevers of the University Medical Center Utrecht in the Netherlands has made the most significant individual impact. Most related studies have been published in high-quality journals, revealing that progress in this field is considered very meaningful. Recent articles with strong citation bursts are good indicators of future research topics. Using various analyses, we found changing trends from “methods to build organoids (e.g., “lgr5+ stem cell” and “3D culture”) to “practical applications of organoids” (e.g., “cystic fibrosis” and “Zika virus”). The main hot topics currently are drug screening, disease modeling, personalized medicine/precision medicine, tissue engineering (regenerative medicine), and developmental biology, and they may still be hotspots in the future. However, there are still some limitations, such as the lack of reproducibility, the low level of maturity and function, and the absence of appropriate functional readouts. Possible solutions involve the development of bioengineering strategies to ensure the homogeneity of organoids, the promotion of vascularization, tissue-tissue interactions, and mimicking physiological environments to increase the organoid maturity and function, as well as the development of accurate and versatile functional readouts to provide more information about the functionality of the organoids.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Funding: This work was supported by the National Natural Science Foundation of China (grant No. 81701929, No. 81772104, and No. 81971889); the Natural Science Foundation of Guangdong Province (grant No. 2019A1515012170, No. 2020A1515110037), and the Science and Technology Program of Guangzhou (grant No. 201904010480).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apm.amegroups.com/article/view/10.21037/apm-22-290/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Checkley W, White AC Jr, Jaganath D, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 2015;15:85-94. [Crossref] [PubMed]
- Heo I, Dutta D, Schaefer DA, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 2018;3:814-23. [Crossref] [PubMed]
- Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299-312. [Crossref] [PubMed]
- Sondo E, Caci E, Galietta LJ. The TMEM16A chloride channel as an alternative therapeutic target in cystic fibrosis. Int J Biochem Cell Biol 2014;52:73-6. [Crossref] [PubMed]
- Weren RD, Ligtenberg MJ, Kets CM, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet 2015;47:668-71. [Crossref] [PubMed]
- Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47-54. [Crossref] [PubMed]
- Gabriel E, Ramani A, Karow U, et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 2017;20:397-406.e5. [Crossref] [PubMed]
- Pamarthy S, Sabaawy HE. Patient derived organoids in prostate cancer: improving therapeutic efficacy in precision medicine. Mol Cancer 2021;20:125. [Crossref] [PubMed]
- Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920-6. [Crossref] [PubMed]
- Dekkers JF, Wiegerinck CL, de Jonge HR, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 2013;19:939-45. [Crossref] [PubMed]
- Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176-87. [Crossref] [PubMed]
- Fujii M, Shimokawa M, Date S, et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016;18:827-38. [Crossref] [PubMed]
- Fordham RP, Yui S, Hannan NR, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 2013;13:734-44. [Crossref] [PubMed]
- Hu H, Gehart H, Artegiani B, et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018;175:1591-1606.e19. [Crossref] [PubMed]
- Xu R, Zhou X, Wang S, et al. Tumor organoid models in precision medicine and investigating cancer-stromal interactions. Pharmacol Ther 2021;218:107668. [Crossref] [PubMed]
- Atala A. Tissue engineering of reproductive tissues and organs. Fertil Steril 2012;98:21-9. [Crossref] [PubMed]
- Tran BX, Vu GT, Ha GH, et al. Global Evolution of Research in Artificial Intelligence in Health and Medicine: A Bibliometric Study. J Clin Med 2019;8:360. [Crossref] [PubMed]
- Zhao Y, Zhang X, Song Z, et al. Bibliometric Analysis of ATAC-Seq and Its Use in Cancer Biology via Nucleic Acid Detection. Front Med (Lausanne) 2020;7:584728. [Crossref] [PubMed]
- Cobo MJ, López-Herrera AG, Herrera-Viedma E, et al. Science mapping software tools: Review, analysis, and cooperative study among tools. J Am Soc Inf Sci Technol 2014;62:1382-402. [Crossref]
- Ghorbani F, Feizabadi M, Farzanegan R, et al. An Investigation of Topics and Trends of Tracheal Replacement Studies Using Co-Occurrence Analysis. Tissue Eng Part B Rev 2017;23:118-27. [Crossref] [PubMed]
- Yao L, Hui L, Yang Z, et al. Freshwater microplastics pollution: Detecting and visualizing emerging trends based on Citespace II. Chemosphere 2020;245:125627. [Crossref] [PubMed]
- Ence AK, Cope SR, Holliday EB, et al. Publication Productivity and Experience: Factors Associated with Academic Rank Among Orthopaedic Surgery Faculty in the United States. J Bone Joint Surg Am 2016;98:e41. [Crossref] [PubMed]
- Li F, Li M, Guan P, et al. Mapping publication trends and identifying hot spots of research on Internet health information seeking behavior: a quantitative and co-word biclustering analysis. J Med Internet Res 2015;17:e81. [Crossref] [PubMed]
- Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373-9. [Crossref] [PubMed]
- Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011;141:1762-72. [Crossref] [PubMed]
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125. [Crossref] [PubMed]
- Clevers H. Modeling Development and Disease with Organoids. Cell 2016;165:1586-97. [Crossref] [PubMed]
- Qian X, Nguyen HN, Song MM, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016;165:1238-54. [Crossref] [PubMed]
- van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933-45. [Crossref] [PubMed]
- Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324-38. [Crossref] [PubMed]
- Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013;13:653-8. [Crossref] [PubMed]
- Garcez PP, Loiola EC, Madeiro da Costa R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016;352:816-8. [Crossref] [PubMed]
- Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262-5. [Crossref] [PubMed]
- Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105-9. [Crossref] [PubMed]
- Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481-4. [Crossref] [PubMed]
- Ke L, Lu C, Shen R, et al. Knowledge Mapping of Drug-Induced Liver Injury: A Scientometric Investigation (2010-2019). Front Pharmacol 2020;11:842. [Crossref] [PubMed]
- Dekkers JF, Berkers G, Kruisselbrink E, et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med 2016;8:344ra84. [Crossref] [PubMed]
- Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc 2020;15:3380-409. [Crossref] [PubMed]
- Boretto M, Maenhoudt N, Luo X, et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol 2019;21:1041-51. [Crossref] [PubMed]
- Jung P, Sato T, Merlos-Suárez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med 2011;17:1225-7. [Crossref] [PubMed]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014;9:2329-40. [Crossref] [PubMed]
- Li H, Saucedo-Cuevas L, Shresta S, et al. The Neurobiology of Zika Virus. Neuron 2016;92:949-58. [Crossref] [PubMed]
- Pauli C, Hopkins BD, Prandi D, et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov 2017;7:462-77. [Crossref] [PubMed]
- Seppälä TT, Zimmerman JW, Sereni E, et al. Patient-derived Organoid Pharmacotyping is a Clinically Tractable Strategy for Precision Medicine in Pancreatic Cancer. Ann Surg 2020;272:427-35. [Crossref] [PubMed]
- Nikolaev M, Mitrofanova O, Broguiere N, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020;585:574-8. [Crossref] [PubMed]
- Lukonin I, Serra D, Challet Meylan L, et al. Phenotypic landscape of intestinal organoid regeneration. Nature 2020;586:275-80. [Crossref] [PubMed]
- Sorrentino G, Rezakhani S, Yildiz E, et al. Mechano-modulatory synthetic niches for liver organoid derivation. Nat Commun 2020;11:3416. [Crossref] [PubMed]
- McCracken KW, Catá EM, Crawford CM, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014;516:400-4. [Crossref] [PubMed]
- Dye BR, Hill DR, Ferguson MA, et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 2015;4:e05098. [Crossref] [PubMed]
- Noguchi TK, Ninomiya N, Sekine M, et al. Generation of stomach tissue from mouse embryonic stem cells. Nat Cell Biol 2015;17:984-93. [Crossref] [PubMed]
- McCracken KW, Aihara E, Martin B, et al. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 2017;541:182-7. [Crossref] [PubMed]
- Volpato V, Smith J, Sandor C, et al. Reproducibility of Molecular Phenotypes after Long-Term Differentiation to Human iPSC-Derived Neurons: A Multi-Site Omics Study. Stem Cell Reports 2018;11:897-911. [Crossref] [PubMed]
- Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun 2012;3:792. [Crossref] [PubMed]
- Martyn I, Kanno TY, Ruzo A, et al. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 2018;558:132-5. [Crossref] [PubMed]
- Lancaster MA, Corsini NS, Wolfinger S, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 2017;35:659-66. [Crossref] [PubMed]
- Uzel SG, Amadi OC, Pearl TM, et al. Simultaneous or Sequential Orthogonal Gradient Formation in a 3D Cell Culture Microfluidic Platform. Small 2016;12:612-22. [Crossref] [PubMed]
- Bagley JA, Reumann D, Bian S, et al. Fused cerebral organoids model interactions between brain regions. Nat Methods 2017;14:743-51. [Crossref] [PubMed]
- Mansour AA, Gonçalves JT, Bloyd CW, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 2018;36:432-41. [Crossref] [PubMed]
- Workman MJ, Mahe MM, Trisno S, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017;23:49-59. [Crossref] [PubMed]
- Koike H, Iwasawa K, Ouchi R, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 2019;574:112-6. [Crossref] [PubMed]
- Okkelman IA, Foley T, Papkovsky DB, et al. Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials 2017;146:86-96. [Crossref] [PubMed]
- Kratz SRA, Höll G, Schuller P, et al. Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems. Biosensors (Basel) 2019;9:110. [Crossref] [PubMed]
- Wang Y, Wang H, Deng P, et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 2018;18:3606-16. [Crossref] [PubMed]


