
The clinical significance of the SIRT2 expression level in the early stage of sepsis patients
Introduction
Sepsis is defined as life-threatening organ dysfunction or tissue hypoperfusion caused by a deregulated host response to infection (1). It is not only the most significant cause of morbidity and mortality in intensive care units (ICU) worldwide but it is also a recognized global health burden (2-4). A recent assessment of global incidence and mortality of hospital-treated sepsis shows that there maybe 31.5 million sepsis cases with potentially 5.3 million deaths annually (5). Early identification and rapid initiation of therapy are recognized as efficacious in reducing the morbidity and mortality of sepsis. Currently, the diagnosis of sepsis almost relies on syndromic-based diagnosis. Biomarkers may be a way to achieve a rapid diagnosis of sepsis in the emergency department. Regrettably, there are no biomarkers that are universally accepted for sepsis diagnosis at present (6).
Sirtuins, a highly conserved family of proteins, are reported to take part in the metabolic and immune response of cells (7). In recent years, researchers have reported that sirtuin2 (SIRT2), 1 of the 7 members of the sirtuin family, is associated with infection and inflammation. One study showed that the expression level of SIRT2 was reduced at the initial stage of lipopolysaccharide (LPS)-induced inflammation in the mouse brain, and SIRT2 could suppress the inflammatory responses in collagen-induced arthritis (8,9). Apart from these, several reports have shown that SIRT2 is related to inflammation in sepsis, and protects against the development of inflammatory processes via increasing NF-κB deacetylation, modulating macrophage polarization, and increasing the M2-associated anti-inflammatory pathway (10). SIRT2 can inhibit the inflammation by deacetylation of NF-Κb/p65 and prevent it from entering the nucleus (11). SIRT2 deficiency can prolong the survival of mice with chronic staphylococcal infection by increasing the bacterial phagocytosis of macrophages (12). Also, SIRT2 has been reported to have the ability to adjust microvascular inflammation in sepsis mice model notably in obese mice with sepsis (13,14). Based on these findings, we can surmise that the expression level of SIRT2 in the sepsis patients may be different to that of healthy persons, and this highly conserved gene might be a potential reference indicator for improving the clinical diagnosis of sepsis. This study therefore investigated the expression level of SIRT2 in sepsis and evaluated its value in assessing sepsis diagnosis through a comparison with procalcitonin (PCT) and hypersensitive C-reactive protein (hs-CRP).
Methods
Study design
This was observational and has been registered in the Chinese Clinical Trial database (ChiCTR1800019038). Following informed consent, 38 critically ill patients, consisting of 22 sepsis and 16 septic shock patients, who were treated at the ICU of Tianjin First Center Hospital between July 2018 and May 2019, were recruited within 24 h of entry into the ICU. The critically ill patients were delineated according to the new definitions for sepsis and septic shock published at The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) (1). Sepsis was defined as life-threatening organ dysfunction caused by a dysregulated host response to infection and septic shock was identified as a clinical construct of sepsis with persisting hypotension requiring vasopressors to maintain mean arterial pressure (MAP) ≥65 mmHg and having a serum lactate level >2 mmol/L despite adequate volume resuscitation (1). The control group included 15 healthy volunteers (9 females and 6 males) with a mean age of 61.8, who were recruited from the physical examination department. All patient samples were treated in accordance with the Declaration of Helsinki (as revised in 2013), and the study was approved by the independent ethics committees at Tianjin First Center Hospital (No. 2018N085KY).
The exclusion criteria included patients younger than 18 years old, severe traumatic brain injury, spinal cord injury resulting in permanent sensory and motor deficits, sepsis with an uncontrollable source (e.g., unresectable bowel ischemia), heart failure, human immunodeficiency virus infection, autoimmune diseases or acquired immune deficiency syndrome, patients who had undergone transplantation, chemotherapy or radiotherapy within 30 days prior to onset of sepsis, expected lifespan <3 months due to severe pre-existing comorbidities, and pregnancy.
Data collection and blood sampling
The following routine inspection data for all patients were recorded: gender; age; source of infection on the admission; the APACHE II score; Sequential Organ Failure Assessment (SOFA) score; PCT and hs-CRP levels on admission, and on the second and fourth study day. The peripheral blood samples were collected on admission, and on the second and fourth study day. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation over Ficoll-Hypaque gradients and prepared for subsequent RNA extraction.
RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction (PCR)
Total RNA was extracted from 2×106 PBMCs using TRIzol reagent (Invitrogen, Grand Island, NY, USA), and then the reverse transcription (RT) reactions were carried out using the SuperScript II RT (Invitrogen) following the manufacturer’s protocol. To detect the miRNA expression of SIRT2 in PBMCs of sepsis patients, we performed real-time quantitative PCR with SYBR-Green PCR kit (Takara, Shiga, Japan) on the ABI PRISM-7500 Sequence detection system (Applied Biosystems, Foster City, CA, USA). The sequences of primers were as follows: for SIRT2: 5'-ACGCTGTCGCAGAGTCAT-3' (forward), 5'-CGCTCCAGG GTATCTATGTT-3' (reverse). Human GAPDH primers used as an internal control were 5'-GAAGG TGAAGG TCGG AGTC-3' (forward) and 5'-GAAGATGG TGATGGG ATTTC-3' (reverse). For PCR amplification, PCR reactions were performed in a total volume of 20 µL, containing 2 µL of sample cDNA, 0.2 µM of each primer and the SYBR-Green PCR kit following the manufacturer’s instructions. Thermal cycling conditions were 95 °C for 30 sec, followed by 40 cycles of 5 sec at 95 °C, and 34 sec at 60 °C. The relative gene expression level of SIRT2 was obtained by comparing the expression of SIRT2 to that of GAPDH using the 2−ΔΔCt method [ΔΔCt = (CtSIRT2 − CtGAPDH) sample − (CtSIRT2 − CtGAPDH) calibrator].
Statistical analysis
The results were presented as median and range as well as mean ± SD. The quantitative data were analyzed with Student’s t-test using Statistical Package for the Social Sciences version 19.0 (IBM Corporation, Armonk, NY, USA). P<0.05 was considered a statistically significant value.
Results
In this study, we recruited 38 sepsis patients from July 2018 and May 2019, including 22 sepsis patients and 16 septic shock patients. There were no significant differences in baseline characteristics in terms of sex, age, and site of infection. The clinical patient status was assessed with the APACHE II score (15), the patients of sepsis group APACHE II score on the first day was 14.1, and the septic shock group’s score was 15.6. The extent of multiple organ failure was evaluated with SOFA (16). The SOFA score on the first day in the sepsis group was 6.5, and it was 9.8 for the septic shock group (Table 1).
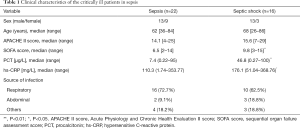
Full table
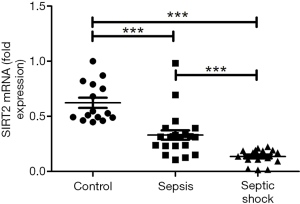
The expression of SIRT2 mRNA in sepsis patients and septic shock patients was lower than that of the healthy volunteers (control group) on the first day (0.331 and 0.136 vs. 0.623; P<0.001), and the expression level of SIRT2 mRNA in septic shock patients was lower than that in the sepsis patients (0.331 vs. 0.136; P<0.001) (Figure 1). Also, the SIRT2 mRNA expression levels in the sepsis and septic shock patients on the second day and fourth day were all lower than those of the SIRT2 mRNA in the healthy volunteers. The mean expression level of SIRT2 mRNA in sepsis patients was 0.165 on the second day and 0.226 on the fourth day. The mean SIRT2 mRNA level in septic shock patients was 0.221 on the 2nd day and 0.217 on the 4th day (Figure 2). The results of both the sepsis group and the septic shock group compared with the control group were significantly different with P<0.001. On the first day, the septic shock patients showed lower expression of SIRT2 mRNA than the sepsis patients (P<0.001), but there was no significant difference on the following days (Figure 2).

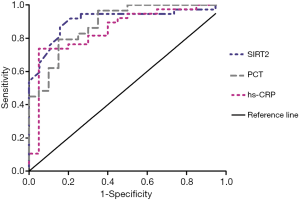
To evaluate the usefulness of SIRT2 expression level on the diagnosis of sepsis, we compared the predictive usefulness of SIRT2 expression level, PCT, and hs-CRP in patients with sepsis by using receiver operating characteristic (ROC) curves (Figure 3). In the ROC curves for sepsis, the area under the curve (AUC) of SIRT2 mRNA expression was larger than the AUC of PCT and CRP. The sensitivity of SIRT2 was higher than the sensitivity of PCT and hs-CRP, and the specificity of SIRT2 was higher than the specificity of hs-CRP but lower than that of PCT in the ROC curves for sepsis. The cut-off value for SIRT2 was 0.393, the AUC of SIRT2 was 0.908, the sensitivity was 0.919, and the specificity was 0.789 (P<0.0001; Youden index =0.708. The cut-off value for PCT was 0.73 µg/L, the AUC of PCT was 0.886, the sensitivity was 0.793, and specificity was 0.85 (P<0.0001; Youden index =0.643. The cut-off value for hs-CRP was 62.54 mg/L, the AUC of hs-CRP was 0.851, the sensitivity was 0.737, and the specificity was 0.75 (P<0.0001; Youden index =0.687) (Figure 3).
Discussion
Sepsis is caused by a dysregulated host response to infection which can include bacterial, fungal, and viral infections, making it difficult to diagnose clinically (6). At present, the diagnosis of sepsis still relies on pyrexia, white blood cell (WBC) counts, heart skip speed, clinical indications, and on culture-based pathogen detection. Even though the “Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic shock” recommends that using the SOFA and Quick SOFA (qSOFA) systems to diagnose sepsis (17,18), these systems are known to lack sensitivity and are not sufficient for accurately diagnosing sepsis. The microbiologic culture results are the gold standard for sepsis diagnosis. However, this examination often needs a few days to return results, and it typically lags days behind the first treatment (17,19,20).
A biomarker is any substance, structure, or process that can be measured in the body or its products. Biomarkers influence or predict the incidence rate or outcome of a disease, and these biomarkers are being researched for use in the diagnosis of sepsis (21,22). A biomarker may provide an essential diagnostic tool for sepsis at an early stage (23). CRP and PCT, with the exception of lactate, are probably the 2 most widely used biomarkers used clinically for diagnostic of sepsis (24-26), but neither of these are universally accepted for sepsis diagnosis (27,28). CRP is a well-established biomarker for infection and inflammation but lacks specificity. It is also elevated in patients with cancer, burns, and other inflammatory conditions but not infectious diseases (28,29). PCT is a precursor-hormone of calcitonin and has been recommended as a useful test in critically ill patients (30). However, much of the literature does not fully consider PCT to be able to accurately distinguish sepsis from systemic inflammatory response syndrome (SIRS) in critically ill patients, as it may be elevated in the absence of infection (28,31,32).
SIRT2, one of the members of the sirtuin family of proteins, is related to infection, DNA damage, cell survival, and many other processes. In recent years, some research has found that it may be associated with inflammation. Some inflammatory factors, such as LPS, can reduce the expression level of SIRT2 (8,9). Regrettably, the mechanisms of this phenomenon are still unclear. Besides this, the current study on the relationship between SIRT2 and inflammation is limited to animal models. Given these reports, we surmise that the expression level of SIRT2 in sepsis patients may be inconsistent with that of a healthy person and might be a potential biomarker for the diagnosis of sepsis. Therefore, we conducted this clinical trial and found that the septic shock patients show a lower expression level of SIRT2 than the sepsis patients on the first day, but there were no significant differences on the following days. The focus of these results was that both the expression levels of SIRT2 in the sepsis and septic shock patients were lower than those of the healthy volunteers.
Moreover, the lower expression level lasted within 4 days from admission. We compared SIRT2 with PCT and hs-CRP via the ROC curve for evaluating the usefulness of SIRT2 in the diagnosis of sepsis. ROC curves showed that SIRT2 has a higher AUC and sensitivity than PCT and hs-CRP. Furthermore, the ROC curve also showed that the specificity of SIRT2 was higher than that of hs-CRP but lower than that of PCT. These suggest that SIRT2 has potential as a diagnostic marker for sepsis.
In the early stages of infection, the body needs to induce an inflammatory response to resist foreign infection, and SIRT2 can inhibit NF-κB single pathway, thus inhibiting the production of inflammatory factors by deacetylating NF-κB/p65 and modulating the polarization of macrophages. The reason for the reduce of SIRT2 expression level in the early stages of sepsis may be due to its protective role during the inflammatory processes. However, it is worth noting that knockdown of the SIRT2 gene instead could protect mice from sepsis in animal experiments. Additionally, there have been reports indicating that the expression of SIRT2 shows an upward trend with the prolongation of LPS stimulation time (8). These results imply that the expression level of SIRT2 might change during the progress of sepsis. Here, we detected the expression levels of SIRT2 in the early stage of sepsis and found that the SIRT2 expression is lower than that in the healthy body. This is preliminary confirmation of SIRT2 as a new biomarker for the early diagnosis of sepsis compared with PCT and hs-CRP. However, there needs to be further verification for its application in the clinical setting. The relationship between SIRT2 and sepsis also needs further research. To find the truth, we will continue to expand case samples and extend observation time in our next study.
Acknowledgments
Funding: This work was supported by the Science and Technology Foundation of Tianjin First Central Hospital [grant numbers: YCM201809] and the National Clinical Key Specially Project Foundation of the Ministry of Health in China [grant numbers: 2011-873].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-19-653). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patient samples were treated in accordance with the Declaration of Helsinki (as revised in 2013), and the study was approved by the independent ethics committees at Tianjin First Center Hospital (No. 2018N085KY).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Mierzchala-Pasierb M, Krzystek-Korpacka M, Lesnik P, et al. Interleukin-18 serum levels in sepsis: Correlation with disease severity and inflammatory markers. Cytokine 2019;120:22-7. [Crossref] [PubMed]
- Kodan LR, Verschueren KJC, Kanhai HHH, et al. The golden hour of sepsis: An in-depth analysis of sepsis-related maternal mortality in middle-income country Suriname. PLoS One 2018;13:e0200281. [Crossref] [PubMed]
- Rudd KE, Kissoon N, Limmathurotsakul D, et al. The global burden of sepsis: barriers and potential solutions. Crit Care 2018;22:232. [Crossref] [PubMed]
- Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med 2016;193:259-72. [Crossref] [PubMed]
- Al Jalbout N, Troncoso R Jr, Evans JD, et al. Biomarkers and Molecular Diagnostics for Early Detection and Targeted Management of Sepsis and Septic Shock in the Emergency Department. J Appl Lab Med 2019;3:724-9. [Crossref] [PubMed]
- Vachharajani VT, Liu T, Wang X, et al. Sirtuins Link Inflammation and Metabolism. J Immunol Res 2016;2016:8167273. [Crossref] [PubMed]
- Pais TF, Szegő ÉM, Marques O, et al. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J 2013;32:2603-16. [Crossref] [PubMed]
- Lin J, Sun B, Jiang C, et al. Sirt2 suppresses inflammatory responses in collagen-induced arthritis. Biochem Biophys Res Commun 2013;441:897-903. [Crossref] [PubMed]
- Lo Sasso G, Menzies KJ, Mottis A, et al. SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS One 2014;9:e103573. [Crossref] [PubMed]
- Wang X, Buechler NL, Long DL, et al. Cysteine thiol oxidation on SIRT2 regulates inflammation in obese mice with sepsis. Inflammation 2019;42:156-69. [Crossref] [PubMed]
- Ciarlo E, Heinonen T, Théroude C, et al. Sirtuin 2 Deficiency Increases Bacterial Phagocytosis by Macrophages and Protects from Chronic Staphylococcal Infection Front Immunol 2017;8:1037. [J]. [Crossref] [PubMed]
- Buechler N, Wang X, Yoza BK, et al. Sirtuin 2 Regulates Microvascular Inflammation during Sepsis. J Immunol Res 2017;2017:2648946. [Crossref] [PubMed]
- Wang X, Buechler NL, Martin A, et al. Sirtuin-2 Regulates Sepsis Inflammation in ob/ob Mice. PLoS One 2016;11:e0160431. [Crossref] [PubMed]
- Knaus WA, Draper EA, Wagner DP, et al. APACHE II: a severity of disease classification system. Crit Care Med 1985;13:818-29. [Crossref] [PubMed]
- Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine Intensive Care Med 1996;22:707-10. [Crossref] [PubMed]
- Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock:2016. Intensive Care Med 2017;43:304-77. [Crossref] [PubMed]
- Teparrukkul P, Hantrakun V, Imwong M, et al. Utility of qSOFA and modified SOFA in severe malaria presenting as sepsis. PLoS One 2019;14:e0223457. [Crossref] [PubMed]
- Lippi G. Sepsis biomarkers: past, present and future. Clin Chem Lab Med 2019;57:1281-3. [Crossref] [PubMed]
- van Engelen TSR, Wiersinga WJ, Scicluna BP, et al. Biomarkers in Sepsis. Crit Care Clin 2018;34:139-52. [Crossref] [PubMed]
- Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS 2010;5:463-6. [Crossref] [PubMed]
- Wong HR. Sepsis Biomarkers. J Pediatr Intensive Care 2019;8:11-6. [Crossref] [PubMed]
- Samraj RS, Zingarelli B, Wong HR. Role of biomarkers in sepsis care. Shock 2013;40:358-65. [Crossref] [PubMed]
- Póvoa P. C-reactive protein: a valuable marker of sepsis. Intensive Care Med 2002;28:235-43. [Crossref] [PubMed]
- Sharma S, Duggal N. Role of procalcitonin, Il-6 and C- reactive protein in suspected cases of sepsis. Indian J Pathol Microbiol 2019;62:578-81. [Crossref] [PubMed]
- Faix JD. Biomarkers of sepsis. Crit Rev Clin Lab Sci 2013;50:23-36. [Crossref] [PubMed]
- Hayashi N, Yamaguchi S, Rodenburg F, et al. Multiple biomarkers of sepsis identified by novel time-lapse proteomics of patient serum. PLoS One 2019;14:e0222403. [Crossref] [PubMed]
- Mierzchała-Pasierb M, Lipińska-Gediga M. Sepsis diagnosis and monitoring - procalcitonin as standard, but what next? Anaesthesiol Intensive Ther 2019;51:299-305. [PubMed]
- Pepys MB, Hirschfield GM. C-reactive protein: A critical update. J Clin Invest 2003;111:1805-12. [Crossref] [PubMed]
- O’Grady NP, Barie PS, Bartlett JG, et al. Guidelines for evaluation of new fever in critically ill adult patients: 2008 update from the American College of Critical Care Medicine and the Infectious Diseases Society of America. Crit Care Med 2008;36:1330-49. [Crossref] [PubMed]
- Reinhart K, Brunkhorst FM. Meta-analysis of procalcitonin for sepsis detection. Lancet Infect Dis 2007;7:500-2. [Crossref] [PubMed]
- Becker KL, Snider R, Nylen ES. Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit Care Med 2008;36:941-52. [Crossref] [PubMed]


