
DNA methylation changes in the hippocampus of learning and memory disorder offspring rats of lead exposure during pregnant and lactation period
Introduction
The hippocampus and cortex are major components of the brain of humans and other vertebrates. Exploring the cellular and molecular mechanisms of memory formation and maintenance has become one of the key hotspots in neuroscience. Researchers have gradually realized the importance of gene transcription and protein synthesis in the formation of long-term memory through various experiments (1). Previous studies have used high-throughput screening technology to search for genes that are expressed in the cortex and hippocampus. Currently, a number of important genes have been reported, and Cheng et al. (2) reviewed these genes. Nearly 100 genes associated with learning and memory have their functions fall in these five categories: synaptic function, signal transduction, energy metabolism, transcriptional and translational regulation, and cell apoptosis.
Although the study of the cellular and molecular mechanisms of learning and memory has been the focus of neuroscience for some time, the epigenetic mechanism behind the dynamic changes of gene transcription responsible for memory formation and maintenance has gradually become the focus of attention in recent years. Epigenetic gene regulation often involves chemical change (physical marking) of DNA or related proteins to cause or allow long-lasting modifications in gene activity (3-5). Epigenetic mechanisms, such as histone modifications (methylation, acetylation, and deacetylation) and DNA methylation, play important roles in learning and memory (6). The relationship between DNA methylation and learning and memory has attracted increasing attention (7,8). Recent studies have confirmed that DNA methylation and the change of its extent may have important influences in the regulation of neurogenesis in aging animals, especially in terms of the learning and memory functions (9,10). Some studies have reflected that neuronal functions and DNA methylation are correlated: BDNF can affect neural plasticity, thereby affecting the functions of learning and memory. Miller et al. further explored the relationship between memory formation and DNA methylation (11).
Epigenetic mechanisms play essential roles in the development and maintenance of addiction as well as memory formation in the brain (12-14). The neurotoxicity of arsenic to children is a serious health problem. The pathogenic mechanism is considered to be the epigenetic change of DNA. Male Sprague-Dawley (SD) rats were treated with arsenic trioxide via drinking water for 6 months. Arsenic exposure impaired the cognitive abilities of these rats, reduced the 5-mC (5-methylcytosine) and 5-hmC (5-hydroxymethylcytosine) levels, and down-regulated DNA methyltransferases (DNMTs) (15). Lead (Pb)impairs children’s cognitive function. Zhang et al. reported the effect of lead acetate on learning and memory ability in rats (16). Exposure to lead (Pb) at concentrations lower than the threshold proposed by the CDC and Prevention (17) is associated with unusual behaviors, learning disabilities and impaired cognitive functions in humans and laboratory animals (18). Yang et al. further showed that Lead contact during pregnancy can result in long-term learning/memory impairments in young adults. Thus, it is natural to question how the molecular mechanisms underlying the learning ability and memory defects in young offsprings after maternal lead exposure work and whether they are involved in epigenetic changes. We present the following article in accordance with the ARRIVE Reporting Checklist (available at http://dx.doi.org/10.21037/apm-19-421).
Methods
Materials
SD rats were acquired from the Hunan Slac-Jingda Experimental Animal Center (Hunan, China). All rats were fed at will. The environmental conditions of the feeding room are as follow: temperature is controlled at 22±2 °C, humidity is controlled at 55%±5%. Light and darkness are given for 12 hours. During the whole experiment, rats were kept in captivity in large plastic cages containing sterile corn cobs (locally obtained) as cushions. In this experiment, a barrier housing facility was used in accordance with the National Standard Laboratory Animal Requirements of Environment and Housing Facilities (GB14925-2010). The care of the laboratory animals and animal experimental operation were performed in accordance with the committee of Jiangxi University of Traditional Chinese Medicine. This study approved by the Animal Care and Research Committee of Jiangxi University of Traditional Chinese Medicine ethics committee (No.2018jz07). No contaminants, which would have interfered with or affected the results of the study, were present in the feed or water. Lead acetate (batch number: 160525) was purchased from Shanghai Haiqu chemical Co., Ltd. All other chemicals were of reagent grade.
Animal experimental design
After 5–7 days of adaptive feeding, 2 females:1 male was paired in the same cage, and the vaginal plug was detected as the 0th day of conception, and then the pregnant rats were randomly divided into a negative control group and a test group (n=9). The test group was treated with lead acetate at the doses of 1 g/L through drinking water from the 1st day pregnancy to the 21th day weaning after birth of their offsprings (total poisoned for 42 days). A separate negative control group was exposed to saline instead of lead acetate. At the time of weaning (21 days after birth), 50 pupils were randomly selected from each group for Morris water maze test to test learning and memory ability and 3 male pupils were randomly selected from each group with remaining weaning rats for examination on DNA methylation changes. The contents of lead in whole blood and hippocampus were detected in 20 pupils in each group. Detailed experimental grouping and treatments are illustrated by Figure 1. After the experiment, the rats were anesthetized by an overdose of sodium pentobarbital and sacrificed.
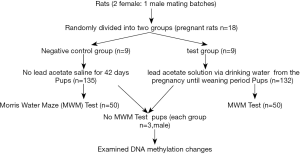
Determination of lead content in blood and hippocampus
Nitrification was performed on newly collected blood and hippocampus samples. Lead level in whole blood and hippocampus was evaluated by graphite furnace atomic absorption spectrometry (GFAAS).
Learning and memory test
Rat learning and memory tests were accomplished as described earlier (19). A black round pool with a diameter of 1 meter and depth of 30 cm was divided into four quadrants, I, II, III, and IV. A platform with a diameter of 11 cm was fixed to the first quadrant, and the pool quadrants were filled with water to 1 cm below the platform. The water temperature was controlled at 22–25 °C. The test data of place navigation and space exploration test (spatial probe) are analyzed using ANOVA.
DNA methylation changes in the hippocampus
Genomic DNA extraction and fragmentation
Genomic DNA was extracted from tissue samples of three control groups and three test groups using a DNeasy Blood & Tissue Kit (Qiagen, Fremont, CA, USA). Purified DNA was then quantified using a nanodrop ND-2000 (Thermo scientific, Wilmington, DE, USA). Genomic DNA from each sample was sonicated to ~200–1,000 bp with a Bioruptor sonicator. Quality analysis of gDNA and fragments of DNA were performed with agarose electrophoresis.
Immunoprecipitation
DNA was heat-denatured at 94 °C for 10 min, then cooled rapidly on ice, immunoprecipitated with 1 µL of the primary antibody overnight at 4 °C with rocking agitation in 400 µL of PBS (0.5% BSA). In order to recover more of the immunoprecipitated DNA fragments, 200 µL of anti-mouse IgG magnetic beads were added, and cultured at 4 °C for 2 hours with stirring. After the above procedure, immunoprecipitation washing was carried out using ice-cold immunoprecipitation buffer for 5 times. The washed beads were resuspended in TE buffer with 0.25% sodium dodecyl sulfate and 0.25 mg/mL proteinase K for 2 hours at 65 °C and then cooled to room temperature. The medip DNA was purified using a Qiagen-mine-lute column (Qiagen, Fremont, CA, USA).
Whole genome amplification (WGA)
MeDIP-enriched DNA was amplified using a WGA kit from Sigma-Aldrich Whole Genome Amplification. The amplified DNA were purified using QIA quick PCR purification kit (Qiagen, Fremont, CA, USA).
DNA labeling and array hybridization
Nanodrop ND-2000 for quantified purified DNA. NimbleGen Dual-Color DNA Labeling Kit was used for DNA labeling. A total of 1 µg of DNA from each sample was incubated for 10 min at 98 °C with 1 OD of the IP sample (Cy5-9 mer primer) or Input sample (Cy3-9mer primer). Then, 100 U of the Klenow fragment (New England Biolabs, USA) and 100 pmol of deoxynucleoside triphosphates were added, and the mixture was incubated at 37 °C for 2 h. Adding 0.1 volume of 0.5 M EDTA to stopped the reaction, and purified labeled DNA with isopropanol/ethanol precipitation. Microarrays were hybridized at 42 °C for 16 to 20 h with Cy3/5 labeled DNA in Nimblegen hybridization buffer/hybridization component A in a hybridization chamber (Hybridization System-Nimblegen Systems, Inc., Madison, WI, USA). After hybridization, Nimblegen Wash Buffer kit was used for washing (Nimblegen Systems, Inc., Madison, WI, USA). A Roche NimbleGen’s Rat DNA Methylation 385K Promoter Plus CpG Island Array was used for array hybridization.
Quality assessment of the raw data
Plotting distribution of the red/green intensity ratio (‘M’) with reference to the average intensity (‘A’) is a MA-plot. The following equations were used to defined M and A. The original signal strengths of the probes for MeDIP and input channels are denoted by R and G, respectively.
[1]
[2]
A correlation matrix described the correlation among replicate experiments, and a Correlation Matrix was used to correlate the raw log2- (IP/Input) values in replicates in R.
Data normalization
We normalized the log2 ratio of raw data values to avoid technical variations and assess differences in methylation amongst samples. We conducted median centering, quantile normalization, and linear smoothing. In this process, we used the bioconductor encapsulation of Ringo, Limma, and Medme methods. After normalization, we created normalized log2 ratio data for each sample for further peak finding analysis.
Peak-finding and methylation enrichment
The Medip chip data was analyzed using a sliding window (750 bp) provided by the roche nimblegen nimblescan version 2.6 on the normalized log2 ratio data using a peak finding algorithm. A one-sided KS (Kolm gorov-Smirnov) test was used to determine whether the plumbs were drawn from a significantly more positive distribution of the intensity log2-ratios than those in the rest of the array. Each plumb received a -log10 P value score from the KS test around that plumb.
The experimental methylation process of the chip detection is shown in Figure 2.
Results
Blood and brain lead level of 21-day-old offsprings after pregnant and lactation period exposure
The scales of lead in serum and hippocampus of offspring in intrauterine and lactation lead exposure groups were 0.291±0.085 and 0.302±0.062 mg/L respectively, and 0.009±0.001 and 0.007±0.001 mg/L in control group, with significant difference (P<0.05).
Learning and memory ability tests in rats
Results of place navigation for 21-day-old young rats
The experimental results of place navigation of 21-day-old pupils of pregnant female rats exposed to lead are shown in Figure 3. The 21-day-old pupils of pregnant and lactating rats exposed to lead demonstrated a significantly affected navigation ability. With the increase of training time, the escape time of the control group was significantly reduced, while the escape time of the 21-day-old offspring rats that were exposed to lead during the pregnancy and lactation period did not decrease significantly, and the difference was significant.

Experimental results of space exploration of 20-day-old young rats
After 7 days of training, the space exploration test was carried out. The time spent searching for the platform in control group was faster than that in the experimental group, and the number of pupils that crossed the desk was also significantly higher than that in the experimental group. There were more differences regarding the time spent in the first quadrant for the experimental group (Figure 4).
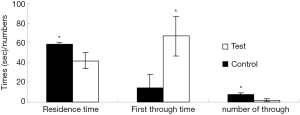
The results of the integrated navigation and space exploration test showed that the learning and memory abilities of 20-day-old pupils of pregnant and lactating rats exposed to lead were illustrious lower than those of rats in the group of control (no lead exposure led to normal young rats).
DNA methylation changes
Quality evaluation of the raw data
Immunoprecipitation experiments compared MeDIP (enriched samples) to Input (control samples) on the same arrange. The M-values are not prospective to center around zero. Correlation and Matrix Data Normalization is shown in Figure 3. Box plots are an easy way to quickly envisage the assignment of a dataset. After normalization, all arrays were centered at the same level to zero, and were equivalently assigned to the log2-ratios across every slide (Figure 5).
Methylation enrichment and peak-finding
A one-sided KS test was used to determine whether the plumbs were drawn from a significantly more positive distribution of intensity log2-ratios than those in the rest of the array. Each probe received a -log10 P value score from the KS test around that probe (*_pvalues.gff).
CpG island methylation and MeDIP array data analysis results
The union between promoter methylation and transcriptional inhibition of downstream genes has been established, and mammalian gene promoters are known to contain divergent GC contents with different methylation profiles. Based on the CpG ratio, Gene promoters are subdivided into three classes according to GC content and length of the CpG-rich region as following: high (denoted HCP); low (LCP); and intermediate (ICP).
The results from the enriched peaks between the lead exposure group and control group on chromosomes 1, 2, 7, and 11 showed that they were still methylated in the HCP region. The degree of DNA methylation of 1 and 11 increased, while that of 2 and 7 displayed no marked change. DNA methylation of chromosomes 5, 9, 18, 19, 20 and 21 of the control group disappeared after lead exposure. However, by contrast, DNA methylation of chromosomes 6, 8, 10 and 17 was found only the experimental group exposed to lead (Figure 6A).

In the group of control, the original chromosome 3 in the ICP region remained methylated, while methylation of chromosomes 8, 17, and 19 disappeared. After exposure to lead, DNA methylation was found at the ICP region of chromosomes 1, 4, 5, 9, 10, 11, and 15 (Figure 6B).
Enrichment peaks from the LCP results between the lead exposure group and the control group were found in the control group on chromosomes 1 and 7. The LCP on chromosome 10 showed slight methylation. Lead exposure significantly improved methylation of chromosome 1 LCP, but the DNA methylation region of chromosome 7 disappeared and the basic methylation of LCP on chromosome 10 showed no change in this region. Moreover, the methylation level of LCP on chromosome 4 in the lead exposed group was relatively high, while the extension of LCP on chromosomes 8, 15, 16, and 21 of the control group exhibited a general low DNA methylation (Figure 6C).
Gene Ontology (GO) analysis
The results of the GO analysis were sorted according to P values. The results were listed according to the following three aspects: BP, CC and MF. In our study, genes found in GO terms, such as for metal ion transport, cell connections, the lamellar body, the axon bulge, and increased methylation of various metal transporters, were significantly enriched (Figure 7).

Pathways analysis
In our study, the hedgehog signaling pathways, neuroactive receptor-ligand interacting ligase pathways, and high methylation pathways that interact with cytokines were identified, as shown in Figure 8.

Discussion
DNA methylation is a genomic biomarker that is susceptible to chemical and environmental factors. Heavy metals are chemicals associated with many diseases, such as cancer, cardiovascular disease, neurological disorders and autoimmune diseases. These metals widely exists in our living environment and have become nonnegligible pollutants. Recently, the importance of the roles that these molecular factors play in the etiology of heavy metal-associated diseases was described. Studies have shown that exposure to toxic metals (including arsenic, cadmium and lead) can lead to changes in DNA methylation (20-23). Growing evidence have reflected that exposure to arsenic (As) can alter DNA methylation and histone modifications (24). DNA hypomethylation in brain tissue of polar bears and mouse ESC (embryonic stem cells) is thought to be associated with mercury exposure (25,26). DNA methylation can be a biomarker of past exposure to lead. The decrease of LINE-1 methylation level is associated with lead levels in patella and tibia (27). Reverse correlation between lead accumulation and genomic DNA methylation levels was reported (28). A series of animal studies have shown that lead exposure is associated with neurotoxicity and attention deficit disorder through epigenetics (29,30).
Epidemiological and tentative studies have found that some environmental events during fetal development have a lasting impact on cell function, thus affecting the track of health events throughout life (31). Maternal exposure to lead can lead to genetic changes in epigenetics, such as changes in DNA methylation, which can affect grandchildren (32).
So far, there is not much data on the effect of lead exposure in the maternal uterus on DNA methylation levels in descendants with reduced learning and memory abilities. In our study, we found that maternal Pb exposure caused deficits of the learning and memory ability of offspring and that the DNA methylation level in the experimental group at the HCP, ICP, and LCP in the chromosome CPG region was higher than that in the control group via methylation chip analysis. GO analysis revealed that the methylation levels of genes related to BP, CC, and MF in the experimental group were lower than those in the control group.
Conclusions
Lead contact during fetal period affects learning and memory ability after birth. The mechanism of lead-induced learning and memory impairment may involve epigenetic changes through DNA methylation.
Acknowledgments
Funding: This work was supported by the grant from the Natural Science Foundation of Jiangxi province (Project No. 20151BAB205064). ZYL was supported this funding. ZYL and LL are responsible for the conception and design of this project.
Footnote
Reporting Checklist: The authors have completed the ARRIVE Reporting Checklist. Available at http://dx.doi.org/10.21037/apm-19-421
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-19-421
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-19-421). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The care of the laboratory animals and animal experimental operation were performed in accordance with the committee of Jiangxi University of Traditional Chinese Medicine. This study was approved by the Animal Care and Research Committee of Jiangxi University of Traditional Chinese Medicine ethics committee (No.2018jz07).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ugajin A, Uchiyama H, Miyata T, et al. Identification and initial characterization of novel neural immediate early genes possibly differentially contributing to foraging-related learning and memory processes in the honeybee. Insect Mol Biol 2018;27:154-65. [Crossref] [PubMed]
- Xiaorui C, Wenxia Z, Zhang Y. Advances in study on learning and memory related genes. Letts In Biotechnol 2003;14:227-30.
- de Meireles LC, Galvão F, Walker DM, et al. Exercise Modalities Improve Aversive Memory and Survival Rate in Aged Rats: Role of Hippocampal Epigenetic Modifications. Mol Neurobiol 2019;56:8408-19. [Crossref] [PubMed]
- Guan JS, Xie H, Ding X. The role of epigenetic regulation in learning and memory. Exp Neurol 2015;268:30-6. [Crossref] [PubMed]
- Kim S, Kaang BK. Epigenetic regulation and chromatin remodeling in learning and memory. Exp Mol Med 2017;49:e281. [Crossref] [PubMed]
- Kim S, Kaang BK. Epigenetic regulation and chromatin remodeling in learning and memory. Exp Mol Med 2017;49:e281. [Crossref] [PubMed]
- Morris MJ, Monteggia LM. Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues Clin Neurosci 2014;16:359-71. [Crossref] [PubMed]
- Day JJ, Sweatt JD. Cognitive neuroepigenetics: a role for epigenetic mechanisms in learning and memory. Neurobiol Learn Mem 2011;96:2-12. [Crossref] [PubMed]
- Deibel SH, Zelinski EL, Keeley RJ, et al. Epigenetic alterations in the suprachiasmatic nucleus and hippocampus contribute to age-related cognitive decline. Oncotarget 2015;6:23181-203. [Crossref] [PubMed]
- Navehbenjamin M, Ohta N. Mem Aging. 2016.
- Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci 2010;13:664. [Crossref] [PubMed]
- Akbarabadi A, Niknamfar S, Vousooghi N, et al. Effect of rat parental morphine exposure on passive avoidance memory and morphine conditioned place preference in male offspring. Physiol Behav 2018;184:143-9. [Crossref] [PubMed]
- Kim HD, Call T, Magazu S, et al. Drug Addiction and Histone Code Alterations. Neuroepigenomics in Aging and Disease. Springer, 2017:127-43.
- Coppedè F. Epigenetics and cognitive disorders—translational aspects. Neuropsychiatric Disorders and Epigenetics. Elsevier, 2017:69-91.
- Du X, Tian M, Wang X, et al. Cortex and hippocampus DNA epigenetic response to a long-term arsenic exposure via drinking water. Environ Pollut 2018;234:590-600. [Crossref] [PubMed]
- Zhang J, Cai T, Zhao F, et al. The role of α-synuclein and tau hyperphosphorylation-mediated autophagy and apoptosis in lead-induced learning and memory injury. Int J Biol Sci 2012;8:935. [Crossref] [PubMed]
- Roeo L. Preventing Lead Poisoning in Young Children. 1991.
- Soleimani E, Goudarzi I, Abrari K, et al. Maternal administration of melatonin prevents spatial learning and memory deficits induced by developmental ethanol and lead co-exposure. Physiol Behav 2017;173:200-8. [Crossref] [PubMed]
- Hamilton DA, Driscoll I, Sutherland RJ. Human place learning in a virtual Morris water task: some important constraints on the flexibility of place navigation. Behav Brain Res 2002;129:159-70. [Crossref] [PubMed]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol 2008;21:28-44. [Crossref] [PubMed]
- Bolin CM, Basha R, Cox D, et al. Exposure to lead and the developmental origin of oxidative DNA damage in the aging brain. FASEB J 2006;20:788-90. [Crossref] [PubMed]
- Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun 2007;352:188-92. [Crossref] [PubMed]
- Takiguchi M, Achanzar WE, Qu W, et al. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res 2003;286:355-65. [Crossref] [PubMed]
- Majumdar S, Chanda S, Ganguli B, et al. Arsenic exposure induces genomic hypermethylation. Environ Toxicol 2010;25:315-8. [Crossref] [PubMed]
- Pilsner JR, Lazarus AL. Mercury‐associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol 2010;19:307-14. [Crossref] [PubMed]
- Arai Y, Ohgane J, Yagi S, et al. Epigenetic assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev 2011;57:507-17. [Crossref] [PubMed]
- Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 2010;118:790. [Crossref] [PubMed]
- Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 2009;117:1466. [Crossref] [PubMed]
- Faulk C, Barks A, Liu K, et al. Early-life lead exposure results in dose-and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 2013;5:487-500. [Crossref] [PubMed]
- Li Y, Xie C, Murphy SK, et al. Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ Health Perspect 2016;124:666. [Crossref] [PubMed]
- Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr 2001;4:611-24. [Crossref] [PubMed]
- Sen A, Heredia N, Senut MC, et al. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci Rep 2015;5:14466. [Crossref] [PubMed]




