
Serum protein complex profiling reveals heterogeneity of Balanced constitutional population in traditional Chinese medicine through blue native polyacrylamide gel electrophoresis
Introduction
The individual differences across people is a topic that has continually attracted interest in clinical medicine. Researchers have put great effort into exploring how modern evolutionary psychology and emotional regulation impacts individual differences and personality (1,2). When it comes to traditional or modern Chinese medicine, it is critical to assess the human constitution in order to distinguish individual differences (3).
Traditional Chinese medicine (TCM) constitution refers to stable morphological features, physiological functions, and psychological states, which emerge from the dichotomy of nature vs. nurture lifestyle influences, and can be conceptually compared to the concept of personalized medicine (4). TCM constitution theory classifies individual constitutions into nine types based on the Chinese medical theoretical framework, with eight of these being unbalanced constitutional types. The theory is used to guide the prevention, diagnosis, and treatment of diseases, along with rehabilitation and health. The Classification and Determination of Constitution in TCM, published by the China Association of TCM, was established by TCM physicians after 30 years of research, and has been tested by clinical experts and epidemiologists. It is an interdisciplinary approach that integrates epidemiology, immunology, molecular biology, genetics, and mathematical statistics into its analysis (5). According to TCM constitution theory, strategies for the prevention and treatment of disease should be always based on the individual’s constitution, and the eight unbalanced constitutions are usually more vulnerable to disease and require special attention. Conversely, the medical attention given to people with Balanced constitution is often lacking, as Balanced constitution does not necessarily ensure an exemption from all sicknesses (6-10).
Indeed, Balanced constitution does not automatically equate to a perfectly healthy condition. When people with Balanced constitution fall sick, the disease will develop according to the characteristics of their constitution (11). In previous studies of the relationship between TCM constitutions and various diseases, such as tumors (12), metabolic diseases (13,14), immune disorders, and emotional disorders (15,16), people with the balanced constitution have accounted for a certain proportion of those with these diseases. In TCM acupuncture treatment, there are obvious differences in the sensation propagated along the meridians among people with different constitutions (17,18). From a clinical perspective, many people with Balanced constitution are in a suboptimal state of health, and specific medical intervention is needed. The individual differences and the heterogeneity of the population may help to explain the occurrence of diseases in the segment of the population with Balanced constitution.
Blood can be a window through which we can peer into a person’s health or illness. Blood analysis is commonly performed to detect disease and determine if the organs are functional (19). In TCM theory, blood is the basis of physiological activities, and thus also fundamentally informs pulse diagnosis. The biological mechanism behind pulse diagnosis has been verified by modern medicine (20,21). Serum contains a variety of proteins that respond to the pathogenesis of disease, which makes it an engaging subject for clinical research (22). The rapid development of serum proteomics has led to the discovery of more single-molecular biomarkers; however, the diagnostic accuracy of these markers is not yet satisfactory (23). As the cornerstone of biological processes, protein complexes perform a vast array of biological functions (24), and the proteins that compose those complexes could serve as indicators of physical status. Blue native polyacrylamide gel electrophoresis (BN-PAGE) combined with mass spectrometry-based proteomic technologies can be used to isolate and detect native proteins and to identify physiological protein-protein interactions in serum (25). This method can also remove some highly abundant proteins from the serum. It may be worthwhile to explore the biological basis of TCM constitution classification by establishing the serum protein profile of different TCM constitutions.
As a cutting-edge technology for TCM classification, proteomics was used to explore the characteristics of individuals with Balanced constitution (26,27). In our study, the protein profiles of 25 healthy volunteers with balanced constitutions were classified into two major patterns. Furthermore, the two patterns of the balanced constitution were found to be associated with TCM unbalanced constitution convert scores, suggesting a transition from Balanced constitution to an unbalanced constitution. Our results represent a valuable approach for investigating individual differences and provide new insights into the categories of TCM classification.
Methods
Serum samples
Volunteers for our study underwent a comprehensive health examination at the Guangdong Provincial Traditional Chinese Medical Hospital, and serum samples were collected afterwards. All of the participants met the classification criteria of the balanced constitution with no experience of disease or medical treatment records within 2 months before their visit. The body constitution status of all the participants was evaluated using the TCM Physical Constitution Scale (4). The main method of measurement was a scientific symptom questionnaire. It is worth noting that all the volunteers came from the same village in Foshan City of the Guangdong Province. They shared a homogeneous genetic background, living environment, and eating habits, which made them an ideal population for this study.
The blood samples from these volunteers were also subjected to blood biochemistry measurements. The serum samples were managed by the Biological Resource Center of the Guangdong Provincial Hospital of Chinese Medicine. The study was approved by the Animal Ethics Committee of the Guangdong Provincial Hospital of Chinese Medicine (B2017-150-01) and was performed in accordance with the tenets of the Declaration of Helsinki (as revised in 2013). Informed consent was given by all participants.
Classification of TCM constitution type
The constitution type of participants was evaluated using the TCM Constitution Scale (4). The survey consists of 60 questions, and there are 5 options for each question (not all, few, sometimes, often, always), corresponding to 1–5 points. There were nine subscales used to assess the nine TCM constitution types individually, including balanced constitution, qi-deficiency constitution, yang-deficiency constitution, yin-deficiency constitution, phlegm-damp constitution, damp-heat constitution, blood-stagnation constitution, qi-stagnation constitution, and inherited special constitution. Firs, we calculated the score by totaling the scores of each subscale. Then, the constitution convert scores of each subscale were calculated by applying the following formula: (original score – the number of questions)/(the number of questions×4) ×100. If the constitution convert scores of the balanced constitution was >60 or the constitution convert scores of the eight unbalanced constitutions were <40 points, Balanced constitution was claimed. If the score of any constitution subscale was ≥40 points, they were deemed to have unbalanced constitutions.
BN-PAGE analysis of the serum sample
Serum protein complexes were resolved and displayed by BN-PAGE as described previously, with some modification (25). Briefly, 2.5 µL of serum sample was mixed with 7.5 µL of sample loading buffer (31.25 mM Tris-HCl, 12.5% glycerol, 0.5% bromophenol blue. pH 7.5). The samples were run on a 3.2% stacking gel followed by 5–11% (w/v) (acrylamide/bisacrylamide =37:1) gradient acrylamide gel [150 mM Bis-Tris, 200 mM 6-Amino acetic acid (pH 7.4)] containing 70% glycerol to separate the protein complexes in the serum. Electrophoresis was performed at 100 V for 1 h at room temperature, followed by 300 V for 7 h at 4 °C. After electrophoresis, the gel was stained with Coomassie Brilliant Blue G-250. The image of the gel was scanned with an HP Scanjet G4050 (Hewlett-Packard, Inc., Palo Alto, CA, USA). The bands of the serum protein complexes were identified by ImageJ software (National Institutes of Health, USA; https://imagej.nih.gov/ij/download.html).
Protein complex pattern analysis
The same amount of prestained molecular weight markers was used as a reference for each individual gel. Gels were manually aligned based on the molecular weight of the markers. Each individual band was labeled with a number based on its molecular weight from the top to the bottom. ImageJ was used for image processing; the gel image was converted into an array of pixels, in which the intensity of the band was represented by peaks. The abundance value of a band was calculated by the sum of areas under the peak. The relative abundance and distinguishable bands were selected by cluster analysis to represent the patterns of protein complexes from each individual serum sample. Consensus clustering was performed with MeV (Multi Experiment View) software, using the hierarchical clustering method with Pearson’s correlation coefficient.
MS sample preparation
The distinguishable representative gel bands were sliced from the BN-PAGE gels for in-gel trypsin digestion as described previously (28). The tryptic peptides were dried under a vacuum for further analysis.
Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis
Peptides were analyzed using an ultra-performance LC-MS/MS platform of a hybrid LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Inc., San Jose, CA, USA) equipped with a Waters nanoACQUITY ultra-performance liquid chromatography system (Waters Corporation, Inc., Milford, MA, USA). Each sample was reconstituted with 10 µL of 0.1% (v/v) formic acid and 1% (v/v) acetonitrile in water for subsequent analysis. The liquid chromatography separation was performed on an in-house packed capillary column (75 µm inner diameter ×15 cm length) with 3-µm C18 reverse-phase fused silica column (Michrom Bioresources, Inc., Auburn, CA, USA). The samples were loaded into the column by an auto-sampler and eluted with a 30-min gradient covering 0–100% of phase B (phase B, 0.1% FA in ACN; phase A, 0.1% FA +2% ACN in water) with a flow rate of 0.3 µL/min. Eluting peptides were analyzed using an LTQ-Orbitrap Velos mass spectrometer. The full scan was performed within m/z 300–1,600 with a resolution of 30,000 at m/z 400. The automatic gain control was set to 1×106, and the maximum injection time was 150 ms. After a full scan, the top 20 most abundant ions were selected for MS/MS analysis under 35% normalized collision energy (NCE) in the linear ion trap (LTQ). The precursor isolation window was set to 2, and the dynamic exclusion time was set to 35 seconds.
Bioinformatics analysis of identified proteins of gel bands
The acquired raw files were searched by MaxQuant software suite (version 1.5.6.0, Martinsried, Germany) against the UniProt human reference protein database (version March, 2017). MS search parameters consisted of full tryptic restriction, fixed modification of cysteine carbamidomethylation, and variable modifications of methionine oxidation. The maximum false discovery rates (FDR) for peptide and protein identification were specified as 0.01. Gene ontology (GO) annotation and pathway analysis were performed using the DAVID website (version 6.8, https://david.ncifcrf.gov/home.jsp) (29,30).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (v6.01, GraphPad software, Inc., San Diego, CA, USA) and SPSS V26.0 (IBM Corporation, Armonk, NY, USA). Variables accounted for in all our analyses. Group-wise comparisons done with chi-square test for categorical variables and ANOVA test for discrete variables. P values less than 0.05 were considered significant. Figures were arranged for publishing using Photoshop CS6 (Adobe Systems, Inc., San Jose, CA, USA).
Results
High reproducibility of human serum protein complex patterns resolved by blue native PAGE
BN-PAGE was applied to detect the functional protein complex in serum samples. To measure the reproducibility of our BN-PAGE, protein electrophoresis was carried out with four technical replicates. Coomassie blue staining showed multiple distinguishable gel bands on each technical replica, ranging from 146 to 1,048 kDa (Figure 1A). All of the bands in the same lane with varied molecular weight and intensity formed specific protein complexes that might have represented the identity of the individual serum sample. The patterns of the bands from all the four replicates were almost identical, indicating the high reproducibility of our BN-PAGE.
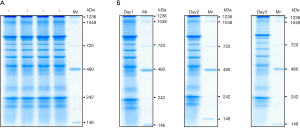
To further verify the reproducibility of BN-PAGE, the technical replicas were analyzed 3 days in a row. The same distinguishable gel bands, ranging from 146 to 1,048 kDa, were observed on each lane in each run (Figure 1B). The overall intensity of the entire lane on different gels varied because of the different staining and destaining times. However, the relative intensity of the gel bands compared to their own molecular weight markers remained the same, indicating the amount of the samples was equivalent. The gel patterns of the protein complexes from all technical replicates of the human serum samples were almost identical which confirmed the high reproducibility of the BN-PAGE technology.
Varied protein complex patterns identified from the people with balanced constitutions
Pulse diagnosis is a traditional Chinese medical technique used in the evaluation of health conditions and disease diagnosis. To understand the biological basis of this diagnostic technique, BN-PAGE was applied to evaluate the protein complex profiles of serum samples from 25 people with Balanced constitution according to pulse diagnosis.
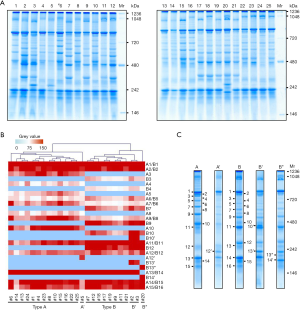
BN-PAGE was used to isolate protein complexes from 25 serum samples (Figure 2A). ImageJ software was used to identify the characteristics of the protein profile. The gel image was converted to gray color in order to be recognized by ImageJ. A marker ladder was used to determine the molecular weight. The number of gel bands was counted, and the intensity of each band was quantified. Additionally, the clustering method was used to identify the protein complex patterns of the 25 samples. The intensity and molecular weight of each gel band were used to generate the cluster (Figure 2B). The results clearly showed two major patterns, type A and type B, and three minor patterns.
In type A, there were 15 bands ranging from 146 to 1,236 kDa, which were labeled A1–A15, respectively. We also noticed a subtype of type A, which was named A’, whose protein complex pattern was very similar to that of type A except there was an additional band located at 242 kDa. This additional band was set as A12’, as its molecular weight was between A12 and A13. There were 16 bands in type B, which were labeled B1–B16. Similarly to type A, type B contained subtypes B’ and B’’. Compared with type B, type B' had two additional bands. B10’ and B13’, while type B’’ had two additional bands (B13’’ and B14’) instead of B14 (Figure 2C).
Notably, band A1-15 appeared in all 14 type A samples, in which 13 samples belonged to type A, and 1 sample belonged to type A’. There were 8, 2, and 1 samples in type B, B’, and B’’, respectively (Figure 2B). The main characteristics of the 25 volunteers in type A and type B are shown in Table S1. No significant differences were found in gender, age, body mass index (BMI), heart rate (HR), systolic blood pressure (SBP), diastolic blood pressure (DBP), fasting plasma glucose (FPG), total cholesterol (TC), total triglyceride (TG), glutamic oxaloacetic transaminase (AST), glutamic-pyruvic transaminase (ALT), blood urea nitrogen (BUN), creatinine (CR), and hypersensitive C-Reactive Protein (hr-CRP).
Enrichment of medium and low abundance serum proteins
As type A and type B were more representative than the other subtypes, one sample from each was selected for mass spectrometry analysis. The number of peptides and peptide-spectrum matches (PSMs) identified in A1–A15 ranged from 429 to 950, and 782 to 1,563, respectively, and the proteins identified by more than two peptides ranged from 48 to 87 in the gel bands of type A and type B (Figure 3A). The sum of the molecular weights of the proteins identified from each band was much larger than the calculated molecular weight of any particular protein complex band, which suggested the presence of multiple protein complexes in each individual band. This was also supported by the phenomena of the molecular weights of the gel bands being disproportionate to the number of identified proteins and peptides. In total, 142 proteins were identified in both the 15 bands of type A and 15 bands of type B (Figure 3B, Figure S1A). In type A, 62 (44%) proteins were immunoglobulin-like proteins, complement-associated proteins, and the proteins which were the top 30 most abundant proteins in normal serum samples. We also identified 80 (56%) proteins with a medium-to-low abundance in the serum (Figure 3C). More interestingly, we noticed that almost one-third of these low abundance proteins were identified in multiple bands (Figure 3D), suggesting medium-to-low abundance proteins in serum were enriched in these gel bands.
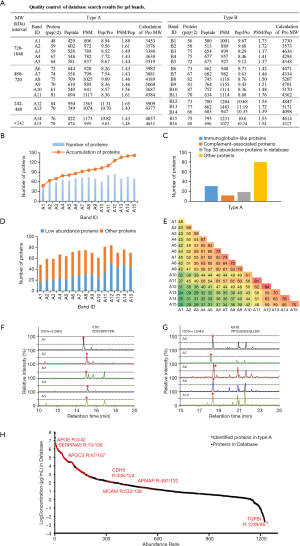
Comparing the protein lists identified from different gel bands, we found that the number of overlapped proteins ranged from 24 to 65, which indicated the difference of these identified protein complexes (Figure 3E). As expected, the abundance of the same protein in different bands was varied. For example, protein vitronectin (VTN), a glycoprotein with high abundance in serum (ranked 30th out of 1,239 in database), was present in A1–A5 with an almost identical peak shape in chromatography and ration time (15 min), and the intensity of VTN was 3.09×105 (100%) which ranked 15th out of 48 in A1 (Table S2). The least abundant protein was 1.10×105 (36%) in A4, and was only one-third of that in A1 (Figure 3F). This suggested that the high abundance proteins in serum were also enriched in some particular bands, but the abundance ranking of the identified proteins in gel bands was far lower than that in the plasma database (ranked 29th out of 1,240 in database). The protein Ig delta chain C region (IGHD) was not included in the serum protein database but was present in A6–A10 at about 17–18 min of ration time. Interestingly, the highest intensity of IGHD was 1.84×105 (100%) in A8. The lowest abundant one was 1.02×105 (55%) in A7, which was only half of that in A8 (Figure 3G). This result clearly showed that the low abundance proteins in the serum samples also accumulated in the protein complex bands. These results further verified that the approach we applied in this study may be helpful in exploring some of the serum proteins that reflected the characteristics of the human population.
We also compared the abundance of the 142 proteins identified from the type A and type B protein complexes with those in the plasma protein database (http://www.plasmaproteomedatabase.org/). Surprisingly, 28 proteins identified in one sample of the type A were not present in the plasma protein database. These newly identified proteins from the isolated protein complexes on the BN gel strongly supported the specificity of the method we applied here. Among the top 200 abundant proteins in the plasma database (31), 94 proteins were also identified from our protein complex samples isolated from the BN gel. The remaining 20 proteins were medium-to-low abundance proteins in serum samples. However, the ranking of the protein abundance in our samples was not consistent with that in the plasma proteome database. For example, apolipoprotein E (APOE), one of the most abundant proteins in the plasma database, was ranked as the 42nd most abundant protein in the 142 proteins identified in type A. Plasma serine protease inhibitor (SERPINA5), which was ranked as the 15th most abundant proteins in the plasma database, was ranked 136th out of 142 in our proteins (Figure 3H). The details of the protein abundance comparison between type B and the plasma database are shown in Figure S1B.
Commonality and specificity of different protein complex patterns at the protein level
About 10 pairs of bands existed in both type A and type B of the balanced constitution samples with the same molecular weight and size, including A1 and B1, A2 and B2, A6 and B5, A7 and B6, A9 and B8, A11 and B11, A12 and B12, A13 and B14, A14 and B15, and A15 and B16 (Figure 2B). The percentage of the overlapped proteins in these paired bands was >58% (Figure 4A). Although some unique proteins might not have existed at the same molecular weight in the paired bands, they were also identified at different molecular weights of the bands. Meanwhile, some unique proteins existed in only one sample but not the other. This further proved the specificity of the band and the difference between type A and type B were due to biological differences. The proteins identified in type A and B serum samples were compared. In total, 124 proteins were found in both of these datasets, with 18 unique proteins present in type A but not in B, and another 18 unique proteins identified in type B but not in type A (Figure 4B). GO analysis showed that the unique proteins in type A were involved mainly in lipid metabolic processes, while the unique proteins in type B were involved in the immune response and inflammatory response pathways (Figure 4C). Cellular components of the unique proteins in type A and type B both existed in extracellular regions (Figure 4D). The molecular function of the unique proteins in type A might be lipoprotein activity, while that of the unique proteins in type B might be endopeptidase activity and antigen binding (Figure 4E).
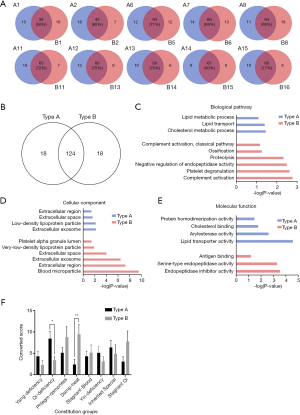
The TCM transformation scores of type A and type B were evaluated using the TCM Constitution Scale. The TCM convert score of qi-deficiency in the type A population was significantly higher than that in the type B population, while the transformation score of damp-heat constitution in the type B population was significantly higher than that in the type A population (Figure 4F). This result indicated that the classification of Balanced constitutional population based on the protein complex pattern may be associated with the transformation score of TCM constitution. There might not be clear-cut boundaries between the human TCM constitution types. People with Balanced constitution of type A may have a tendency towards a qi-deficiency constitution, while people with balanced constitution of type B may share some characteristics of a damp-heat constitution. This study provided us with a reference to reclassify the balanced constitutional population based on the protein profile.
Discussion
TCM constitution should be considered as an approach to describe the state of human health. However, Balanced constitution does not necessarily equate to a healthy state. People with balanced constitutions can also get sick, and the development of disease depends on the particular tendency of each balanced constitution. Therefore, research involving the balanced constitution needs to be taken seriously (32).
The current classification and determination of TCM constitution is mainly determined by a score which is based on the exclusion questions, making the balanced constitution more akin to a hybrid of the constitutions which could not be classified to any other specific unbalanced constitution (33). Our study explored the protein complex patterns of people with a TCM balanced constitution and their associations with unbalanced constitution convert scores. This is the first investigation into the classification of the balanced constitution to study their innate characteristics using serum proteomics.
Some clinical studies have shown that there were significant differences in the pulse patterns among different constitutions and the balanced constitution normally exhibited gentle and slow pulses (34-37). The study of protein complexes in blood can enrich the modern scientific connotation of TCM pulse diagnosis and construct a more complete and objective TCM constitution theory. In the serum proteomics studies conducted by other groups, a commercial affinity removal column was used to remove high abundance proteins from the serum, which is convenient but costly. In another study, serum was denatured by denaturants (38,39), which resulted in the natural state of the serum proteins being insufficiently maintained. In our study, BN-PAGE reduced the percentage of high-abundance proteins but specifically enriched medium-to-low abundance proteins in serum, which was low-cost and easy to operate. The people with balanced constitutions were categorized according to their protein complex patterns. BN-PAGE–based serum protein complex analysis identified two stable and reproducible types of protein complex patterns for the balanced constitution population, which provided a new method for the classification of TCM constitution.
The significance of TCM constitution research is related to preventive treatment, which focuses on adjusting the balance of the immune system and metabolism (40). When changes to the internal and external environment result in imbalance of the immune system or metabolic disorder (41,42), the balanced constitution tends to become unbalanced. People with the qi-deficiency constitution always have abnormal energy metabolism, in which lipid metabolism is important (43,44). In this study, people of the type Balanced constitution had a higher qi-deficiency convert score, and the unique proteins of type A were enriched in the pathways related to lipid metabolism, suggesting that people of type A may be at risk of metabolic diseases. This result is consistent with previous reports that the proportion of obesity, diabetes, and hypertension in the people with qi-deficiency constitutions is higher than that in healthy people (45,46). Damp-heat is a pathogenic factor in traditional Chinese medicine, and is closely related to immune dysfunction of the body (47). The gene expression in people with the damp-heat constitution exhibits an abnormally high immune response, which can manifest as various diseases. The appropriate TCM treatment of this type starts by reducing the body’s inflammatory response and restoring the dynamic balance of the immune system (48-50). The type B population had a higher damp-heat constitution score, and its unique proteins were enriched in immune- and inflammation-related biological pathways, which indicates the association between type B and the damp-heat constitution. Immune regulation, inflammatory response, and lipid metabolism may be the potential mechanisms for the individual differences among people with Balanced constitution.
Conclusions
In summary, BN-PAGE is a robust method to isolate protein complexes in serum, and the use of BN-PAGE followed by LC-MS/MS technology is effective to establish the serum protein complex profile. Our study was the first to provide insight into the association of protein complex patterns with individual differences among the TCM balanced constitution population, and verifies some common characteristics between the balanced and unbalanced constitution types. This novel serum proteomic profile, which used protein complexes as indicators of physical status, can help TCM professionals to identify the characteristics of people with the balanced constitution, thus providing a direction for increasingly targeted therapies for the different TCM constitutions.
Acknowledgments
Funding: This work was supported by the Chinese National Basic Research Programs (no. 2017YFC0906600 and 2017YFD0501500), the Foundation of Guangdong Provincial Hospital of Traditional Chinese Medicine (no. YN2018DB02, YN2018RBA01, and 2016KT1206), the National Natural Science Foundation of China (no. 31670834, 31700723, 31870824, 31901037, and 91839302), the Chinese National Key Project Specialized for Infectious Diseases (no. 2018ZX10302302), the Guangzhou Science and Technology Innovation & Development Project (no. 201802020016), and the Foundation of State Key Lab of Proteomics (no. SKLP-K201704).
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/apm-20-290
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-290
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-290). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Animal Ethics Committee of Guangdong Provincial Hospital of Chinese Medicine (B2017-150-01) and performed in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was given by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Buss DM. How can evolutionary psychology successfully explain personality and individual differences? Perspect Psychol Sci 2009;4:359-66. [Crossref] [PubMed]
- John OP, Gross JJ. Healthy and unhealthy emotion regulation: personality processes, individual differences, and life span development. J Pers 2004;72:1301-33. [Crossref] [PubMed]
- Wang J, Li YS, Ni C, et al. Cognition research and constitutional classification in Chinese medicine. Am J Chin Med 2011;39:651-60. [Crossref] [PubMed]
- Wang Q. Classification and determination of constitution in TCM. Beijing: China Press of Traditional Chinese Medicine, 2009.
- Wang Q. Constitutional doctrine of traditional Chinese medicine. Beijing: People’s Medical Publishing House, 2008.
- Zhang HM, Wang J. Study on the pathogenesis of diseases caused by biased constitution from the gene-environment interaction. China Journal of Traditional Chinese Medicine and Pharmacy 2015;30:3580-2.
- Li LR, Wang J, Li YS, et al. Basic research status of syndromes and diseases of TCM and the necessity of introducing dimension of TCM constitution. China Journal of Traditional Chinese Medicine and Pharmacy 2017;32:2347-9.
- Wang QF, Wang QB, Wang Q. Guidance of TCM constitution theory on disease prevention. Liaoning Journal of Traditional Chinese Medicine 1993;3:15-8.
- Dong LP. Association of TCM constitution theory and disease. Chinese Journal of Basic Medicine in Traditional Chinese Medicine 2006;12:859-60.
- Liu J. The theory of constitution of traditional Chinese medicine and the treatment of diseases. Chinese Journal of Basic Medicine in Traditional Chinese Medicine 2009;15:316-7.
- Zhou LY, Liu J, Zhang JQ, et al. Reflection on the balanced constitution and its identification method. China Medical Herald 2016;13:62-5.
- Dong J, Duan MM, Wang K. Investigation of TCM constitutions and psychological status of tumor patients. Journal of Qilu Nursing 2015;21:53-4.
- Zhai F, Gao H. Application of traditional chinese medicine constitution identification in health management of chronic diabetes patients. J Clin Med Pract 2018;22:62-5.
- Zhu L, Liu LG. Research of traditional Chinese medicine constitutions in diabetes mellitus patients. J Exp Clin Med 2015;14:1311-3.
- Han SH, Kong XD, Song LX, et al. Correlation analysis of traditional Chinese medicine constitution types and rheumatoid arthritis. Liaoning Journal of Traditional Chinese Medicine 2017;19:192-5.
- Yang XY, Zhang LK, Sheng L, et al. Relationship between anxiety, depression, and TCM constitution. Chinese Journal of Integrative Medicine on Cardio- Cerebrovascular Disease 2017;15:1903-1905.
- Yang X, Li XZ, Fu NN, et al. Effect of acupuncture for pain threshold among the groups of different constitutions. Chinese Acupuncture & Moxibustion 2016;36:491-5. [PubMed]
- Wang Q, Tan YQ. Research on relativity of the propagated sensation along meridian and traditional Chinese medicine constitution of middle-aged person. China Journal of Traditional Chinese Medicine and Pharmacy 2017;32:1811-5.
- Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chem Rev 2007;107:3601-20. [Crossref] [PubMed]
- Wu HK, Ko YS, Lind YS, et al. The correlation between pulse diagnosis and constitution identification in traditional Chinese medicine. Complement Ther Med 2017;30:107-12. [Crossref] [PubMed]
- Tsai YN, Chang YH, Huang YC, et al. The use of time-domain analysis on the choice of measurement location for pulse diagnosis research. J Chin Med Assoc 2019;82:78-85. [Crossref] [PubMed]
- Ray S, Reddy PJ, Jain R, et al. Proteomic technologies for the identification of disease biomarkers in serum: advances and challenges ahead. Proteomics 2011;11:2139-61. [Crossref] [PubMed]
- Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA 2011;305:2200-10. [Crossref] [PubMed]
- Hartwell LH, Hopfield JJ, Leibler S, et al. From molecular to modular cell biology. Nature 1999;402:C47-C52. [Crossref] [PubMed]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc 2006;1:418-28. [Crossref] [PubMed]
- Yang J, Yang LC, Li BX, et al. iTRAQ-based proteomics identification of serum biomarkers of two chronic hepatitis B subtypes diagnosed by traditional Chinese medicine. Biomed Res Int 2016;2016:3290260. [Crossref] [PubMed]
- Xu M, Deng JW, Xu KK, et al. In-depth serum proteomics reveals biomarkers of psoriasis severity and response to traditional Chinese medicine. Theranostics 2019;9:2475-88. [Crossref] [PubMed]
- Xu P, Duong DW, Seyfried NT, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009;137:133-45. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002;1:845-67. [Crossref] [PubMed]
- Zhu Y, Shi H, Wang Q, et al. Association between nine types of TCM constitution and five chronic diseases: a correspondence analysis based on a sample of 2,660 participants. Evid Based Complement Alternat Med 2017;2017:9439682. [Crossref] [PubMed]
- Xin H, Wu JK, Guo X, et al. Discussion on the problems of nine traditional Chinese medicine constitutional types scale in clinical application. China Journal of Traditional Chinese Medicine and Pharmacy 2014;29:1841-3.
- Wang Q. Discussion on the methodology of TCM constitution research. Global Traditional Chinese Medicine 2010;3:295-7.
- Zhou GZ, Zhang QC. Investigation of key factors in TCM constitution assessment. Jilin Journal of Traditional Chinese Medicine 2011;31:291-2.
- Liu HH. TCM constitution identification of five elements in TCM. Hubei Journal of Traditional Chinese Medicine 2015;37:36-8.
- Chen YK, Liu Q, Qu HY, et al. Study on characteristics of radial artery pulse diagnositic information among healthy people with Qi-deficiency constitution. Journal of Hunan University of Chinese Medicine 2018;38:289-91.
- Petricoin EF, Liotta LA. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotechnol 2004;15:24-30. [Crossref] [PubMed]
- Cho WC, Yip TT, Yip C, Yip V, Thulasiraman V, Ngan RK, Yip TT, Lau WH, Au JS, Law SC, Cheng WW, Ma VW, et al. Identification of serum amyloid a protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res 2004;10:43-52. [Crossref] [PubMed]
- Cheng N, Li YS, Wang Q. Review and prospect of traditional Chinese medicine constitution research for 40 years. Tianjin Journal of Traditional Chinese Medicine 2019;2:108-11.
- Han XJ, Zhang GQ. Thoughts on the direction of moxibustion development. Journal of Practical Traditional Chinese Internal Medicine 2017;31:69-71.
- Xu QL, Li YF, Jiang SQ, et al. Discussing the following the prognosis of a disease theory application and the problems in modern medicine. Journal of Henan University of Chinese Medicine 2010;25:74-76.
- Bi JL, Yan MH, Chen JY, et al. PIasma metabonomic analysis of healthy qi-deficiency constitution afte rthe intervention by Buzhongyiqi pills. Journal of Chongqing Medical University 2014;38:1124-7.
- Bi JL, Chen JY, Cheng JR, et al. The comparative plasma metabonomic analysis of Qi-deficiency constitution and gentleness constitution. J Trop Med 2014;14:717-20.
- Luo H, Li LR, Wang Q. Correlation between qi deficiency constitution and disease: a bibliometric analysis of 332 observational studies. Tianjin Journal of Traditional Chinese Medicine 2019;36:625-30.
- Yu RX, Li YS, Wang Q. Three problems of traditional Chinese medicine intervention of obese people. China Journal of Traditional Chinese Medicine and Pharmacy 2016;31:3601-3.
- Zhou SS, Lu XJ, Feng LC, et al. Screening of differentially expressed genes and their functions in Damp-heat constitution of Chinese medicine. Liaoning Journal of Traditional Chinese Medicine 2017;44:1819-23.
- Guo JC, Lin J, Xin L, et al. The influence of traditional Chinese medicine for invigorating the spleen on the immune inflammation of rheumatoid arthritis patients with dampness heat Bi obstruction based on association rules. Lishizhen Medicine and Materia Medica Research 2017;28:1002-4.
- Miao B, Wang HQ, Zheng SQ. Effect of changing decoction on immune function, cyto inflammatory factor and intestinal flora in acute ulcerative colitis rats of Damp-heat type. Journal of Emergency in Traditional Chinese Medicine 2019;28:1747-50.
- Zhang NN, Cui C, Zhang ZW. Effect of Zaoshihewei formula on HPA axis, inflammatory factors and immune function in rats with dampness heat syndrome of spleen and stomach. Chinese Journal of Modern Applied Pharmacy 2019;36:1633-8.


