
Macrophage colony-stimulating factor could evaluate both disease activity and renal involvement in systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disorder characterized by autoantibody production and chronic inflammation targeting multiple organs (1,2). The accurate assessment of disease activity in patients with systemic lupus erythematosus (SLE) is a critical tool for clinical practice, observational studies and clinical trials. The SLE disease activity index (SLEDAI), which includes the SLEDAI 2000 (SLEDAI-2K) and Safety of Estrogens in Lupus Erythematosus National Assessment (SELENA)-SLEDAI versions, is the most widely used SLE disease activity measure (3,4). The development of comorbidities and tissue damage, especially renal involvement, can be the direct consequence of SLE or a result of SLE medication (5). Renal involvement is a serious manifestation of systemic lupus erythematosus (SLE), it may portend a poor prognosis as it may lead to end-stage renal disease (6). At present, renal biopsy remains the “gold standard” for the diagnosis of lupus nephritis, but not all patients accept this method because of its invasive nature. However, few reliable biomarkers have been validated for the evaluation of SLE progression (7,8); therefore, there is a need to develop new biomarkers for both disease activity and renal involvement.
In patients with SLE, the immune system is subverted to target numerous self-antigens, and the ensuing inflammatory response elicits a vicious cycle of immune cell activation. Aberrations of monocyte/macrophage phenotype and function are increasingly recognized in patients with SLE and animal models of the disease (9,10). Under physiological conditions, monocytes and macrophages represent an essential arm of the innate immune system with a multitude of immunological functions, including antigen presentation, phagocytosis, and cytokine production. They are heterogeneous cells whose phenotype and functions are regulated by the surrounding micro-environment (11-13). Classically activated or M1 macrophages, which are polarized by interferon-gamma (IFN-γ) or granulocyte-macrophage colony-stimulating factor (GM-CSF), possess high pro-inflammatory, tumoricidal and anti-microbial activities and produce cytokines such as interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α). Alternatively activated or M2 macrophages, which are immune-regulatory and polarized by interleukin-4 (IL-4) and macrophage colony-stimulating factor (M-CSF), are related to the wound healing and anti-inflammatory role and produce cytokines such as interleukin-10 (IL-10) (14,15). Additionally, interleukin-34 (IL-34), sharing the same receptor with M-CSF, is reported to polarize macrophages to an M2-like regulatory phenotype (16,17). Furthermore, the soluble urokinase plasminogen activator receptor (uPAR), expressed on macrophages, is positively related to M2 phenotype macrophages (18,19).
The implications of monocyte/macrophage abnormalities in the pathogenesis of SLE have been discussed in previous studies (20-23). These aberrations include macrophage polarization that is augmented by dysregulated cytokine production (24), and several macrophage-associated cytokines are reported to reach high levels in SLE patients (22,23), e.g., the increased expression of M-CSF, and the correlation between the M-CSF level and SLE activity, are reported in SLE patients (25). However, neither the combined expression of monocyte/macrophage-associated cytokines, nor the direct relationship between monocyte/macrophage and the cytokines, have been sufficiently studied in SLE patients.
In this study, we measured the levels of a panel of 8 cytokines (IFN-γ, GM-CSF, IL-6, TNF-α, M-CSF, IL-34, IL-10, and uPAR) that are characteristic of different macrophage polarization phenotypes. We found that M-CSF was a promising marker to evaluate both disease activity and renal involvement in SLE, with the high level of M-CSF in SLE patients being predominantly derived from monocytes. Our findings emphasize the value of M-CSF in the clinical management of SLE and provide clues regarding key roles of the monocyte/macrophage lineage in SLE progression.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/apm-20-2607).
Methods
Data collection
A total of 100 patients diagnosed with SLE and 44 healthy people were analyzed in Henan Provincial People’s Hospital (Zhengzhou, China). All participants met the 1997 American College of Rheumatology (ACR) revised criteria for the classification of SLE (26); SLE patients with cancer, infectious diseases, and other rheumatic diseases, were excluded. For some analyses, the 100 SLE patients were divided into an inactive group (n=50) and an active group (n=50) according to their SLEDAI score [if ≥10, they were considered clinically active (27)]. For other analyses, these participants were grouped into a non-lupus nephritis group (non-LN, n=52) and lupus nephritis group (LN, n=48) according to the results of renal biopsy in department of pathology in our hospital [including1 class II, 9 class III, 25 class IV, 6 class V, 5 class III + V, and 2 class VI, using the International Society of Nephrology/Renal Pathology Society (ISN/RPS) 2003 lupus nephritis classification (28)]. The pathological activity of lupus nephritis was also evaluated by active index (AI) and chronic index (CI) (29). Demographic information, clinical data, and laboratory findings were extracted from electronic medical records. All participants provided written informed consent, and the research was approved by the Ethics Committee of Henan Provincial People’s Hospital (No: 2019-50). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cytokine detection
Luminex liquid suspension chip technology (Luminex 200, Millipore, Burlington, MA, USA) was used to detect the expression of 8 macrophage-associated cytokines (M1 macrophage: IFN-γ, GM-CSF, IL-6, TNF-α; M2 macrophage: M-CSF, IL-34, IL-10, uPAR) in serum from both SLE and healthy groups. The samples were performed in duplicate according to the manufacturer’s instructions (LXSAHM-08, Research and Diagnostic Systems, Inc., Minneapolis, MN, USA). Data were analyzed using cytokine standard protocols.
Cell sorting
Peripheral blood cells were treated by red blood cell lysis buffer according to standard protocols. The cells were labeled with antibodies to determine their phenotypes (CD15+CD16+ for neutrophils, CD14hi for monocytes, CD3+ CD4+ for CD4 T cells, CD3+ CD8+ for CD8 T cells, and CD20+ B cells), then sorted using Aria III (Becton, Dickinson and Co., Franklin Lakes, NJ, USA) according to standard protocols.
The antibodies used in fluorescence-activated cell sorting (FACS) (Biolegend, USA) were: CD14 (FITC, clone: M5E2), CD16 (PE, clone: 3G8), CD15 (APC, clone: W6D3), CD3 (PE-cy7, clone: HIT3a), CD4 (PE, clone: OKT4), CD8 (FITC, clone: SK1), CD20 (APC, clone: 2H7).
Real-time polymerase chain reaction
Total RNA was extracted from the respective sorted cells using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and reverse transcription was performed using the Super Script First-Strand Synthesis System for real-time polymerase chain reaction (RT-PCR) (Invitrogen, Carlsbad, CA, USA) according to suggested protocols. The RT-PCR was performed using an ABI Prism 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The messenger RNA (mRNA) expression of M-CSF was analyzed by the relative quantification (RQ) value, calculated by normalization to the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by the 2-∆∆Ct method [ΔΔCt = (CtTARGET − CtGAPDH)sample − (CtTARGET − CtGAPDH)calibrator]. The relative expression of M-CSF mRNA in SLE patients was 2-∆∆Ct.
The human primers used in the study were:
GAPDH-Forward (5’-3’) GGAGTCCACTGGCGTCTTCA;
GAPDH-Reverse (5’-3’) TTGCTGATGATCTTGAGGCTGTTG;
M-CSF-Forward (5’-3’) CCAGAAGGAGGACCAGCAAGT;
M-CSF-Reverse (5’-3’) CCAGCAAGACCAGGATGACACT.
Statistical analysis
The software SPSS 17.0 (SPSS, IBM, Chicago, IL, USA) was used for statistical analysis. Categorical variables were presented as n (%), and continuous variables were presented as mean ± SEM, respectively. We used the chi-squared (χ2) test, Fisher’s exact test, unpaired Student’s t-test, and one-way analysis of variance (ANOVA) to evaluate the significance of differences. Detector performance analysis of the cytokine levels for predicting disease activity or renal involvement in SLE was performed using receiver operating characteristic (ROC) curves. The value at which the sum of sensitivity and specificity reached the maximum was considered to be the optimal cutoff according to Youden’s index (J = sensitivity + specificity − 1). Correlation analysis of continuous variables was performed by Pearson correlation analysis. A P value <0.05 was considered statistically significant.
Results
Demographic characteristics and laboratory findings in SLE patients
A total of 100 SLE patients and 44 healthy people were enrolled in this study. To reduce bias, we matched the factors of age and gender upon collection of information and serum from healthy people. Thus, there was no significant difference between healthy controls and SLE patients for age or gender (Table S1).
The 100 SLE patients were divided into an inactive group (n=50) and an active group (n=50) according to the SLEDAI score. There was no significant difference between the two SLE groups regarding age or gender. Additionally, the disease duration of active SLE patients was not different from that of the inactive group. Notably, there were more LN participants in the active group (n=36) than the inactive group (n=12), indicating the close relationship between renal involvement and SLE activity. The features of laboratory findings were also analyzed. The levels of 24-hour urinary protein, serum creatinine (Scr), and C-reactive protein (CRP) and the erythrocyte sedimentation rate (ESR), double-stranded DNA (dsDNA), and complement 3 (C3) were all significantly higher in the active SLE group than in the inactive SLE group (Table S2), suggesting the clinical importance of these laboratory indicators for SLE activity.
The high level of monocyte/macrophage-associated cytokines in SLE patients
In this study, we analyzed the levels of the 8 cytokines related to SLE activity (Figure 1A). Of the 8 cytokines, 6 (all but GM-CSF and IL-34) had higher levels in the active SLE group than in both the inactive SLE group and the healthy control group. The cytokines IFN-γ, IL-6, and TNF-α, are involved in M1 macrophage polarization, while M-CSF, IL-10, and uPAR are involved in M2 macrophage polarization. The results indicated the predictive role of these 6 cytokines for SLE activity.
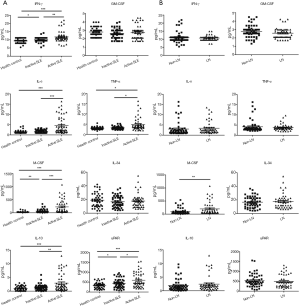
We also analyzed the levels of these 8 cytokines in the non-LN and LN groups to explore their association with renal involvement (Figure 1B). Among the 8 cytokines, only M-CSF had a higher level in the LN group than in the non-LN group. Accordingly, M-CSF can be used to distinguish renal involvement in SLE patients.
The detection efficiency analysis of the cytokine levels for predicting disease activity and renal involvement in SLE patients
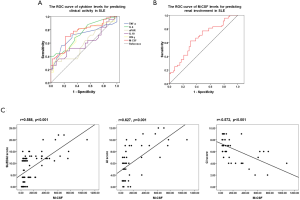
To verify the detection efficiency of these cytokine levels for predicting disease activity in SLE patients, the area under the ROC (AUROC) curve was used to evaluate specificity and sensitivity. Additionally, the optimal cut-off value was calculated according to Youden’s index. The AUROC curves for M-CSF (0.755), IL-6 (0.742), IFN-γ (0.704), and TNF-α (0.702) were significant for predicting disease activity in SLE patients. The optimal cut-off values were 68.750, 3.575, 10.275, and 3.165, respectively (Figure 2A and Table 1), suggesting their utility in predicting SLE activity.

Full table
The detection efficiency of M-CSF level for predicting renal involvement in SLE patients was also evaluated by its ROC curve. The AUROC curve was 0.667 and the optimum cut-off value was 68.750 (Figure 2B), indicating the utility of M-CSF in predicting renal involvement in SLE. These results indicated that M-CSF can predict both disease activity and renal involvement in SLE patients.
The correlation between serum M-CSF level and laboratory or clinical data of SLE patients
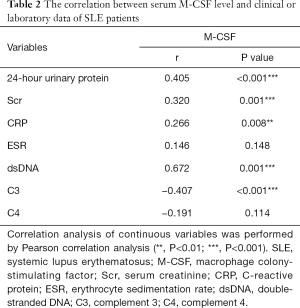
Based on the predictive effect of M-CSF level for both disease activity and renal involvement in SLE patients, the correlation between the serum M-CSF level and laboratory or clinical data was explored to confirm the clinical value of the M-CSF level in SLE (Table 2). For laboratory data, the M-CSF level was positively correlated with the levels of 24 hour urinary protein, Scr, and CRP, dsDNA, and was negatively correlated with the C3 level. For the clinical data, SLE activity was evaluated by SLEDAI scores, and the pathological activity of lupus nephritis was evaluated by renal activity index (AI) and chronicity index (CI) scores (Figure 2C). The results showed that the M-CSF level was positively correlated with the SLEDAI and renal AI scores and negatively correlated with the renal CI scores. This confirmed the potential of M-CSF level to predict both disease activity and renal involvement in SLE patients.
Full table
The high level of M-CSF in SLE patients was closely associated with monocytes
The cytokine M-CSF is reported to be synthesized by mesenchymal cells such as fibroblasts, osteoblasts, and endothelial cells (30). It can also be produced by activated macrophages, B cells, and T cells (31,32). Based on the results above, we identified a close relationship between the serum M-CSF level and cell subpopulations in the peripheral blood of SLE patients. Neutrophils, monocytes, and lymphocytes were quantified in peripheral blood to explore the source of the high level of M-CSF in SLE. The results showed that the numbers of neutrophils and monocytes in SLE patients were significantly higher than in healthy people, indicating that neutrophils and monocytes may contribute to the high level of M-CSF in SLE patients (Figure 3A).
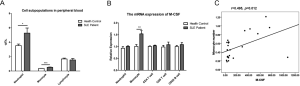
Next, the candidate cells from peripheral blood in both SLE patients and healthy people were sorted (gating strategies in Figure S1) to detect the mRNA expression of M-CSF by RT-PCR. The results showed that the mRNA expression of M-CSF in monocytes from the SLE group was significantly higher than that from the healthy group (Figure 3B). Furthermore, M-CSF level was positively correlated with monocyte numbers (Figure 3C), indicating that the high level of M-CSF in SLE patients was closely associated with monocytes, specifically, the high level of M-CSF in SLE patients was derived mainly from monocytes.
Discussion
The disease SLE is a multisystem autoimmune disorder associated with widespread inflammation and is often progressive. Aberrations of monocyte and macrophage phenotype and function have been increasingly recognized in SLE (33). These aberrations include macrophage polarization augmented by the dysregulated production of cytokines (24,32). Herein, we explored the levels of 8 cytokines associated with macrophage polarization in SLE patients and analyzed the data according to SLE activity or renal involvement. Interestingly, the levels of 4 cytokines (IL-6, IFN-γ, M-CSF, and TNF-α) were related to SLE activity, while only the level of M-CSF was related to renal involvement. In line with other reports (34-36), the predictive role of M-CSF in particular for both SLE activity and renal involvement, cannot be overemphasized.
As an important cytokine for the differentiation and function of the monocyte/macrophage lineage, M-CSF regulates the production and activation phenotype of macrophages (32) and is considered a marker of M2 macrophages (24). Furthermore, M-CSF could be produced by activated macrophages and lymphocytes in addition to mesenchymal cells (37,38). However, unlike M-CSF, GM-CSF plays a minor role in the physiological support of myeloid cells. It is becoming increasingly clear that GM-CSF is a major mediator of tissue inflammation though neutrophils from SLE patients display resistance possibly to the apoptosis-inhibiting effects of GM-CSF (39,40). In this study, the high level of M-CSF, but not GM-CSF, in serum of SLE patients was confirmed to have come mainly from monocytes, which emphasized the involvement of the monocyte/macrophage lineage in SLE progression.
Previous studies have demonstrated that monocyte/macrophage associated cytokines are important participants in SLE. For example, IL-34 has been reported to be correlated with SLE activity (41), the baseline serum levels of IL-6, uPAR and IFN-γ can be used as sensitive biomarkers for disease activity, as well as predictors of remission of lupus nephritis (22,42), the involvement of IL-10 and TNF- genetic variants on SLE appearance, clinical phenotype, and outcome, showed a relevant role of the cytokines in SLE (43). However, neither was correlated with SLE activity or renal involvement in our current study. The differences from previous studies might have been due to the sample size and tissue differences in this research.
There were several limitations to our study. Certain cytokines involved in the study are not specific to macrophages; e.g., IL-6 and TNF-α, are produced by multiple cell types and have a variety of cell targets (24,44-46). Our results showed that 6 of 8 cytokines in a panel had high levels in active SLE patients, indicating that cells of the monocyte/macrophage lineage contribute at least partly to SLE progression. Additionally, drug effects were ignored in this study. The use of anti-inflammatory drugs and immunosuppressants, e.g., methotrexate, may have influenced the levels of these cytokines. Further investigations are needed to evaluate the effects of different drugs.
Conclusions
In summary, our study demonstrated that M-CSF is a promising effective marker to evaluate both disease activity and renal involvement in SLE, providing clues for clinical diagnosis and treatment. The high level of M-CSF in SLE patients was predominantly derived from monocytes, revealing the involvement of the monocyte/macrophage lineage in SLE.
Acknowledgments
Funding: This work was supported by grants 82002210 from the National Natural Science Foundation of China (NSFC).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/apm-20-2607
Data Sharing Statement: Available at http://dx.doi.org/10.21037/apm-20-2607
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/apm-20-2607). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All participants provided written informed consent, and the research was approved by the Ethics Committee of Henan Provincial People’s Hospital (No: 2019-50). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scherlinger M, Mertz P, Sagez F, et al. Worldwide trends in all-cause mortality of auto-immune systemic diseases between 2001 and 2014. Autoimmun Rev 2020;19:102531. [Crossref] [PubMed]
- Teng X, Brown J, Choi SC, et al. Metabolic determinants of lupus pathogenesis. Immunol Rev 2020;295:167-86. [Crossref] [PubMed]
- Kaul A, Gordon C, Crow MK, et al. Systemic lupus erythematosus. Nat Rev Dis Primers 2016;2:16039. [Crossref] [PubMed]
- Jesus D, Matos A, Henriques C, et al. Derivation and validation of the SLE Disease Activity Score (SLE-DAS): a new SLE continuous measure with high sensitivity for changes in disease activity. Ann Rheum Dis 2019;78:365-71. [Crossref] [PubMed]
- Abulaban KM, Song H, Zhang X, et al. Predicting decline of kidney function in lupus nephritis using urine biomarkers. Lupus 2016;25:1012-8. [Crossref] [PubMed]
- Alarcón GS, McGwin G Jr, Petri M, et al. Time to renal disease and end-stage renal disease in PROFILE: a multiethnic lupus cohort. PLoS Med 2006;3:e396. [Crossref] [PubMed]
- Austin HA. Clinical evaluation and monitoring of lupus kidney disease. Lupus 1998;7:618-21. [Crossref] [PubMed]
- Yap DY, Yung S, Ma MK, et al. Serum immunoglobulin G level in patients with lupus nephritis and the effect of treatment with corticosteroids and mycophenolate mofetil. Lupus 2014;23:678-83. [Crossref] [PubMed]
- Liu AC, Yang Y, Li MT, et al. Macrophage activation syndrome in systemic lupus erythematosus: a multicenter, case-control study in China. Clin Rheumatol 2018;37:93-100. [Crossref] [PubMed]
- Ayoub S, Hickey MJ, Morand EF. Mechanisms of disease: macrophage migration inhibitory factor in SLE, RA and atherosclerosis. Nat Clin Pract Rheumatol 2008;4:98-105. [Crossref] [PubMed]
- Orme J, Mohan C. Macrophage subpopulations in systemic lupus erythematosus. Discov Med 2012;13:151-8. [PubMed]
- Burbano C, Villar-Vesga J, Vasquez G, et al. Proinflammatory Differentiation of Macrophages Through Microparticles That Form Immune Complexes Leads to T- and B-Cell Activation in Systemic Autoimmune Diseases. Front Immunol 2019;10:2058. [Crossref] [PubMed]
- Horuluoglu B, Bayik D, Kayraklioglu N, et al. PAM3 supports the generation of M2-like macrophages from lupus patient monocytes and improves disease outcome in murine lupus. J Autoimmun 2019;99:24-32. [Crossref] [PubMed]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 2018;233:6425-40. [Crossref] [PubMed]
- Jaguin M, Houlbert N, Fardel O, et al. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell Immunol 2013;281:51-61. [Crossref] [PubMed]
- Boulakirba S, Pfeifer A, Mhaidly R, et al. IL-34 and CSF-1 display an equivalent macrophage differentiation ability but a different polarization potential. Sci Rep 2018;8:256. [Crossref] [PubMed]
- Zhao Z, Pan G, Tang C, et al. IL-34 Inhibits Acute Rejection of Rat Liver Transplantation by Inducing Kupffer Cell M2 Polarization. Transplantation 2018;102:e265-e274. [Crossref] [PubMed]
- Hu J, Jo M, Eastman BM, et al. uPAR induces expression of transforming growth factor beta and interleukin-4 in cancer cells to promote tumor-permissive conditioning of macrophages. Am J Pathol 2014;184:3384-93. [Crossref] [PubMed]
- Lindsten T, Hedbrant A, Ramberg A, et al. Effect of macrophages on breast cancer cell proliferation, and on expression of hormone receptors, uPAR and HER-2. Int J Oncol 2017;51:104-14. [Crossref] [PubMed]
- Li F, Yang Y, Zhu X, et al. Macrophage Polarization Modulates Development of Systemic Lupus Erythematosus. Cell Physiol Biochem 2015;37:1279-88. [Crossref] [PubMed]
- Li F, Zhu X, Yang Y, et al. TIPE2 Alleviates Systemic Lupus Erythematosus Through Regulating Macrophage Polarization. Cell Physiol Biochem 2016;38:330-9. [Crossref] [PubMed]
- Wen S, He F, Zhu X, et al. IFN-gamma, CXCL16, uPAR: potential biomarkers for systemic lupus erythematosus. Clin Exp Rheumatol 2018;36:36-43. [PubMed]
- Gigante A, Gasperini ML, Afeltra A, et al. Cytokines expression in SLE nephritis. Eur Rev Med Pharmacol Sci 2011;15:15-24. [PubMed]
- Yang X, Feng W, Wang R, et al. Repolarizing heterogeneous leukemia-associated macrophages with more M1 characteristics eliminates their pro-leukemic effects. Oncoimmunology 2017;7:e1412910. [Crossref] [PubMed]
- Yang PT, Xiao WG, Zhao LJ, et al. Increase in the level of macrophage colony-stimulating factor in patients with systemic lupus erythematosus. Ann Rheum Dis 2008;67:429-30. [Crossref] [PubMed]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [Crossref] [PubMed]
- Sayhi S, Achour TB, Mezri S, et al. Clinical Features of Systemic Lupus Erythematosus in Tunisian Males. Curr Rheumatol Rev 2020;16:139-42. [Crossref] [PubMed]
- Weening JJ, D'Agati VD, Schwartz MM, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol 2004;15:241-50. [Crossref] [PubMed]
- Austin HA 3rd, Muenz LR, Joyce KM, et al. Diffuse proliferative lupus nephritis: identification of specific pathologic features affecting renal outcome. Kidney Int 1984;25:689-95. [Crossref] [PubMed]
- Ushach I, Zlotnik A. Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J Leukoc Biol 2016;100:481-9. [Crossref] [PubMed]
- Frétier S, Besse A, Delwail A, et al. Cyclosporin A inhibition of macrophage colony-stimulating factor (M-CSF) production by activated human T lymphocytes. J Leukoc Biol 2002;71:289-94. [PubMed]
- Wang R, Feng W, Yang F, et al. Heterogeneous effects of M-CSF isoforms on the progression of MLL-AF9 leukemia. Immunol Cell Biol 2018;96:190-203. [Crossref] [PubMed]
- Hirose S, Lin Q, Ohtsuji M, et al. Monocyte subsets involved in the development of systemic lupus erythematosus and rheumatoid arthritis. Int Immunol 2019;31:687-96. [Crossref] [PubMed]
- Tian S, Li J, Wang L, et al. Urinary levels of RANTES and M-CSF are predictors of lupus nephritis flare. Inflamm Res 2007;56:304-10. [Crossref] [PubMed]
- Maruyama J, Inokuma S. Cytokine profiles of macrophage activation syndrome associated with rheumatic diseases. J Rheumatol 2010;37:967-73. [Crossref] [PubMed]
- Menke J, Amann K, Cavagna L, et al. Colony-stimulating factor-1: a potential biomarker for lupus nephritis. J Am Soc Nephrol 2015;26:379-89. [Crossref] [PubMed]
- Douglass TG, Driggers L, Zhang JG, et al. Macrophage colony stimulating factor: not just for macrophages anymore! A gateway into complex biologies. Int Immunopharmacol 2008;8:1354-76. [Crossref] [PubMed]
- Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol 2008;8:533-544. [Crossref] [PubMed]
- Becher B, Tugues S, Greter M. GM-CSF. From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 2016;45:963-73. [Crossref] [PubMed]
- Armstrong DJ, Whitehead EM, Crockard AD, et al. Distinctive effects of G-CSF, GM-CSF and TNFalpha on neutrophil apoptosis in systemic lupus erythematosus. Clin Exp Rheumatol 2005;23:152-8. [PubMed]
- Wada Y, Gonzalez-Sanchez HM, Weinmann-Menke J, et al. IL-34-Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Fas(lpr) Mice. J Am Soc Nephrol 2019;30:244-59. [Crossref] [PubMed]
- Abdel Galil SM, Ezzeldin N, El-Boshy ME. The role of serum IL-17 and IL-6 as biomarkers of disease activity and predictors of remission in patients with lupus nephritis. Cytokine 2015;76:280-7. [Crossref] [PubMed]
- López P, Gutierrez C, Suarez A. IL-10 and TNFalpha genotypes in SLE. J Biomed Biotechnol 2010;2010:838390. [Crossref] [PubMed]
- Funes SC, Rios M, Escobar-Vera J, et al. Implications of macrophage polarization in autoimmunity. Immunology 2018;154:186-95. [Crossref] [PubMed]
- Wang LX, Zhang SX, Wu HJ, et al. M2b macrophage polarization and its roles in diseases. J Leukoc Biol 2019;106:345-58. [Crossref] [PubMed]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787-95. [Crossref] [PubMed]
(English Language Editor: J. Jones)


