
Comparing different algorithms for the course of Alzheimer’s disease using machine learning
Introduction
Alzheimer’s disease (AD) is a degenerative disease involving the central system and complex genes (1). The clinical feature of AD is progressive cognitive decline, which leads to the complete need for nursing care within a few years of clinical diagnosis (2). AD is the leading cause of dementia in people aged over 60 years. In 2015, there were 46.8 million dementia patients, and it was estimated that this figure would double every 20 years (3). By 2050, there will be 131.5 million dementia patients, of which 2/3 will be AD patients (3). AD places significant economic and psychological pressures on society and families.
At present, the etiology of AD is not clear. The risk factors of AD are aging, and apoE ε4 gene subtypes, gender, hypercholesterolemia, head trauma, education level, and depression (4). The diagnosis of AD is mainly based on multiple variables and factors, including genetic information, neuropsychological tests, cerebrospinal fluid biomarkers, and brain imaging data (5). Mild cognitive impairment (MCI) occurs in the intermediate state between normal elderly and dementia. MCI is a risk factor of AD, and it can develop into AD (2). MCI is the second stage of AD progression (6). If a patient displays symptoms and pathological changes of cognitive impairment, they are diagnosed with AD. The incidence rate of MCI is 9.9% of the world’s population, and the incidence rate of MCI is 14% to 18% among individuals aged over 70 years (7).
The etiology and pathogenesis of AD remain unclear. At present, the clinical diagnosis mainly depends on the magnetic resonance imaging (MRI) analysis of patients and a neurological scale score that is used to judge their conditions. This method is highly subjective, time-consuming, and laborious, and carries a risk of misdiagnosis (8). The progression of AD is irreversible, but the treatment of MCI in the early stage of AD can delay the passage of the disease. Thus, the question of how to accurately distinguish among normal cognitive (NC), MCI, and AD to implement interventions and adjuvant treatments is essential in clinical settings (9). Feature extraction is of great significance in the clinical diagnosis of AD. Detecting the leading indicators of AD could reduce diagnostic procedures, costs, and time, which would lead to a better classification and prediction effect and provide a reference for clinical diagnosing (10).
Most MRI classification indexes are directly selected based on experience, and they have limited prediction ability and low prediction accuracy. In this study, we did not determine the above lesion sites immediately; rather, we reduced the dimensions of many MRI data. We input the reduced features into random forest (RF), support vector machine (SVM), and decision tree (DT) classifiers and conducted a cross-validation to evaluate the models to select the optimal model to assist in diagnosing the disease. This is an exploration of different machine learning for classify different course of AD. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/apm-21-2013).
Methods
Study design and participants
Based on the AD Neuroimaging Initiative (ADNI) (11), subjects aged between 60 and 90 who had received an education (or participated in work) for at least 6 years and met the diagnostic criteria for each group were selected. The subjects were required to have no contraindications for an MRI examination, were not allowed to participate in other tests, and signed informed consent forms. The inclusion criteria for each group were as follows: (I) cognitive normal (CN) group: match the NC elderly group in terms of age, gender, and education level; show no memory loss (excluding physiological amnesia); have an Mini-Mental State Examination (MMSE) score of 24–30 points; clinical dementia rating (CDR) is 0; have NC function without MCI or dementia; and face no barriers to basic daily activities; (II) early MCI (EMCI) group: be aged from 60 to 90 years; have a junior high school graduation level or above; have a MMSE of 24–30 points; in relation to the logical memory scale, have an education level as follows: ≥16 years (9–11 points), 8–15 years (5–9 points), 0–7 years (3–6 points); have a CDR of 0.5; have no other cognitive impairment; and have no dementia; (III) late MCI (LMCI) group: compared to the EMCI group, the differences were as follows: an education level ≥16 years (≤8 points), 8–15 years (≤4 points), 0–7 years (≤2 points); (IV) AD group: have a MMSE score of 20–26; have a CDR score of 0.5 or 1.0; and be diagnosed with AD according to the new standard of NINCDS/ADRDA (12). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
MRI data preprocessing
First, spatial standardization was used because there are differences in each individual brain and batch processing cannot be used nor can a standard template be used to extract features from each brain scan. To perform the spatial standardization, we used FreeSurfer software (http://surfer.nmr.mgh.harvard.edu) (12). The spatial standardization was performed to eliminate the influence of individual differences. The brain region of all subjects matched the standard brain template (see Figure 1), and the average MRI map collected reflected the template.
We also smoothed the image to reduce noise. As the original image is not very clear, smoothing can improve the clarity of the image. The uniformity of the field intensity affects the gray level, which has a great effect on brain tissue segmentation. It is necessary to correct non-uniform fields. Each image was divided into several sub-images, and half of the adjacent regions overlapped each other. The maximum value of the gray value image was taken as the reference value of the sub-image white matter, and the non-brain tissue signal and noise signal were removed.
We also segmented images with different brain templates, and additional brain tissue features. The gray image was used for pre-segmentation, and the geometric information was then used for fine segmentation. Each part of the image was modulated by different levels of brightness, and the modulated image reflected changes in tissue volume. Using the above process, we identified 272 MRI attributes. There were 69 cortical volume (CV), 49 subcutaneous volume (SV), 68 thickness of cortical area (TA), 70 surface area (SA), and 16 hippocampal subfield (HS) items.
Original MRI features
At the beginning of this study, 272 original MRI features were obtained, comprising 69 CV, 49 SV, 68 TA, 70 SA, and 16 HS items. The unit of CV, SV, and HS is mm3, the division of TA is mm, and the division of SA is mm2.
Machine learning theory
The RF classifier is a combined classifier algorithm composed of many DT classification models (13). The RF classifier extracts multiple samples from the original illustrations using the bootstrap resampling method. First, each piece is modeled by a DT, and then these DT models are gathered together, and the final result is selected by voting. The purpose of the algorithm is to construct a set of tree classifiers and use the collection for classification and prediction by voting. The RF classifier is composed of many DTs. The classification ability of any individual tree may be minimal. However, after many DTs, each tree’s classification results can vote on a test sample to select the best classification model. As the RF machine learning algorithm achieves good performance and perfectly makes up for the low performance of a single classifier, the classifier can be used in various classifications and predictions.
The SVM is a learning method based on the structural risk minimization criterion, which takes the training error as the constraint condition, and the minimum confidence range as the optimization objective (14). The main advantage of this method is that it overcomes the problems of overlearning and falling into a local minimum in traditional ways and has a good generalization ability. A SVM can make a decision function for the linear classification sample set to classify the training set samples correctly, and ensures the classification hyperplane has good generalization ability. The obtained classification hyperplane must have the maximum classification interval. For linear data, the SVM introduces a non-linear mapping in the low dimensional space to the high dimensional space, which is linearly separable in the high dimensional space. The classification learner is then used to solve the problem.
A DT is a classic classification method. It uses a tree structure to make decisions to classify data by rules (14). It is mainly completed by two steps. First, the sample set is trained to generate a DT, and then the DT is pruned. Using a test sample set to verify the rules, any branch that affects the prediction results is cut off. Second, according to the distribution characteristics, the data are divided into different regions to ensure that all the samples contained in the node belong to the same category. The higher the purity, the better.
Statistical analysis
The code and data for the machine learning algorithms designed in this research were all carried out in RStudio software version 1.1.419 (RStudio Inc.). Clear environment variables were defined before each operation and the importation of data. A row represented a sample, and a column represented a feature attribute. The workspace window was mainly used for training and prediction. The main work was carried out in this window, including programming in the editor, debugging the debugging window, and implementing the program running. The results were displayed in the results window. Due to the wide range of values and different units of the original data, the direct input of the data reduced the model’s performance and resulted in the characteristic indexes with large values being assigned a more significant weight. Thus, to ensure the uniformity of data distribution in the measurement space, it was necessary to normalize the data.
The data set was divided into 10 similar mutually exclusive subsets, and each subset was consistent in terms of data distribution. Each time, the union of 9 subsets was used as the training set to build the prediction model, and the remaining subset was used as the test set to test the model’s performance. Thus, 10 training/test sets were obtained, and the average of the 10 results was finally obtained. Training set samples were used to train the data and generate models from the data. Next, the test set samples were input into the model for testing to verify the performance of the selected model. The classification accuracy, specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve (AUC) of the 10-fold cross-validation method was used to evaluate the models. In general, the higher the accuracy, the higher the sensitivity and specificity, and the larger the AUC value, the better the model performance.
Results
Participants
Five hundred and sixty eligible subjects numbered 0–15,000 in the ADNI database were randomly selected. Based on the diagnostic criteria of the ADNI, the subjects were divided into four groups: the CN group (n=140), 230 patients in the EMCI group, 110 patients in the LMCI group, and 80 patients in the AD group.
The data used in this study were all obtained from the ADNI database. The demographic data included data on gender, age, and education level. MMSE ± standard deviation were used to describe each patient’s age, education level, and MMSE score. A ratio was used to describe the sex ratio. The means were evaluated by variance. The non-numerical data were tested by chi-square tests. Indicators including age, education level, and MMSE scores meet the normality and homogeneity of variance. A variance test was carried out, and a chi-square test was carried out for gender. The results (see Table 1) showed that there were no differences in gender and education level among the four groups (P>0.05), but there was a difference in age and MMSE scores (P<0.05).
Table 1
| Variables | CN | EMCI | LMCI | AD | P |
|---|---|---|---|---|---|
| N | 140 | 230 | 110 | 80 | – |
| Gender (male/female) | 73/67 | 126/104 | 74/36 | 40/40 | 0.51 |
| Age | 75.24±6.02 | 72.04±6.53 | 75.53±7.08 | 73.87±6.81 | <0.01 |
| Education | 16.47±2.25 | 16.97±2.51 | 17.11±2.42 | 14.16±0.85 | 0.38 |
| MMSE | 28.43±1.14 | 28.25±1.52 | 23.51±1.75 | 21.09±2.05 | <0.01 |
CN, cognitive normal; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease; MMSE, Mini-Mental State Examination.
Test results
Table 2 sets out the results of the analysis of the collected MRI data. The CVs, SVs, TAs, SAs, and the volume of the HSs gradually decreased over the course of the disease, indicating that patients’ brains slowly atrophied.
Table 2
| MRI | CN | EMCI | LMCI | AD |
|---|---|---|---|---|
| CV (mm3) | 10,132.56±1,162.56 | 9,725.52±1,286.24 | 9,225.84±1,354.48 | 8,215.02±1,562.26 |
| SA (mm2) | 32,542.54±2,562.56 | 29,257.45±2,248.54 | 27,416.85±2,154.24 | 25,846.57±1,884.46 |
| HS (mm3) | 952.05±114.25 | 901.25±119.56 | 854.18±129.57 | 784.43±121.85 |
| SV (mm3) | 1,438.54±179.55 | 1,379.46±220.57 | 1,278.94±214.25 | 1,098.45±154.25 |
| TA (mm) | 3.67±0.35 | 3.67±0.35 | 3.67±0.35 | 3.67±0.35 |
MRI, magnetic resonance imaging; CN, cognitive normal; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease; CV, cortical volume; SA, surface area; HS, hippocampal subfield; SV, subcutaneous volume; TA, thickness of cortical area.
Table 3 shows the comprehensive comparison results of the SVM, RF, and DT classifiers. Combined with specificity, sensitivity, and AUC, the prediction accuracy of the SVM, RF, and DT classifiers was analyzed. As Table 3 shows, in relation the three classifiers, the accuracy of the RF was better than that of the SVM in all AD courses, and the accuracy of the SVM was better than that of the DT. The prediction accuracy of the RF from high to low was CN-AD, EMCI-AD, CN-LMCI, LMCI-AD, EMCI-LMCI, and CN-EMCI. The ranking of the SVM and DT in the six courses was also consistent with that of the RF. In the prediction of CN-AD, the RF’s AUC of 0.92 was higher than that the AUCs of the SVM and DT. The AUCs of the RF for CN-EMCI, CN-LMCI, CN-AD, EMCI-LMCI, EMCI-AD and LMCI-AD were 0.59, 0.81, 0.92, 0.75, 0.85, and 0.89, respectively. The AUCs of the SVM for the six courses were 0.58, 0.68, 0.91, 0.62, 0.71, and 0.54, respectively. The AUCs of the DT for the six courses were 0.57, 0.62, 0.85, 0.58, 0.61, and 0.45, respectively. Thus, among the three prediction schemes, the RF was the best, SVM was the second best, and DT was the worst.
Table 3
| Algorithms | CN-EMCI | CN-LMCI | CN-AD | EMCI-LMCI | EMCI-AD | LMCI-AD |
|---|---|---|---|---|---|---|
| SVM | ||||||
| Accuracy (%) | 65.51 | 74.12 | 90.18 | 70.84 | 86.51 | 72.71 |
| Sensitivity (%) | 78.25 | 61.45 | 81.54 | 29.04 | 68.54 | 58.15 |
| Specificity (%) | 27.58 | 83.15 | 92.41 | 70.18 | 93.75 | 80.45 |
| AUC | 0.58 | 0.68 | 0.91 | 0.62 | 0.71 | 0.54 |
| RF | ||||||
| Accuracy (%) | 77.45 | 87.56 | 96.14 | 81.25 | 90.15 | 84.54 |
| Sensitivity (%) | 79.51 | 64.71 | 88.14 | 38.74 | 93.51 | 67.91 |
| Specificity (%) | 33.54 | 83.94 | 92.81 | 85.94 | 92.43 | 72.46 |
| AUC | 0.59 | 0.81 | 0.92 | 0.75 | 0.85 | 0.89 |
| DT | ||||||
| Accuracy (%) | 61.84 | 71.52 | 87.27 | 65.24 | 81.84 | 69.43 |
| Sensitivity (%) | 75.85 | 58.58 | 78.97 | 27.89 | 67.58 | 56.54 |
| Specificity (%) | 25.85 | 81.45 | 89.57 | 68.74 | 90.41 | 78.14 |
| AUC | 0.57 | 0.62 | 0.85 | 0.58 | 0.61 | 0.45 |
SVM, support vector machine; RF, random forest; DT, decision tree; CN, cognitive normal; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease; AUC, area under the receiver operating characteristic curve.
Figure 2 shows three kinds of machine learning algorithms compared with the original data and the five MRI indexes. The MRI parameters included the 272 raw data, the five features included in the algorithm calculation. (i.e., SV, CV, SA, TA, and HS). The prediction results included CN-EMCI, CN-LMCI, CN-AD, EMCI-LMCI, EMCI-AD, and LMCI-AD.
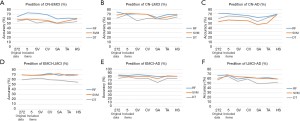
As the five parts of Figure 2 show, the RF had the best prediction effect across all the disease courses. According to the prediction results of CN-EMCI in Figure 2A, in relation to the three machine learning algorithms, the prediction effect of the RF for different MRI features was better than that of the SVM and the DT. The accuracy of the RF, SVM, and DT was 73.8%, 60.7%, and 59.5%, respectively. According to the prediction results of CN-LMCI (see Figure 2B), the highest accuracy of the RF for the 5 included results for the 272 original data was 75.2%, the highest accuracy of the SVM for the five included indexes was 72.4%, and the highest accuracy of the DT for the five indexes was 69%. As Figure 2C shows, the prediction results of CN-AD showed that the highest accuracy of the RF for the 272 original data was 90.5%, the highest accuracy of the SVM for HS was 87.2%, and the highest accuracy of the DT for HS was 87.8%. As Figure 2D shows, the prediction results of EMCI-LMCI showed that the highest accuracy of the RF for CV was 73.5%, the highest accuracy of the SVM for SA was 72.2%, and the highest accuracy of DT for SV was 63.8%. As Figure 2E shows, the prediction results of EMCI-AD showed that the highest accuracy of the RF for SV was 86.5%, the highest accuracy of the SVM for TA was 73.2%, and the highest accuracy of DT for the five included indicators was 80.8%. As Figure 2F shows, the prediction accuracy of LMCI-AD was 69.7% for RF, 68.2% for SVM, and 66.8% for DT.
Discussion
As an important “data pre-processing” process, feature selection can solve dimension disasters in authentic tasks caused by too many attributes. In this study, we identified 272 MRI features. If we had only selected the attributes according to the location of the AD, such as the hippocampus, parahippocampal gyrus, and medial temporal lobe, we would not have been able to find some potential lesions of AD. If only 272 attributes had been analyzed, a dimension disaster would have occurred. By selecting the essential features from them, we ensured that no vital components were lost and the “redundant features” were removed, and thus reduced the difficulty of the machine learning tasks. There were 272 original MRI features, including 69 CV, 49 SV, 68 TA, 70 SA, and 16 HS items. After data dimensionality reduction by statistical methods, a total of five characteristic indexes were identified.
Patients with early stage of AD may have memory impairment, depression, sleep and wakefulness disorders, and sexual dysfunction. The hippocampus is mainly responsible for learning and memory, and its atrophy reflects the severity of AD (15). As MCI progresses to AD, the volume of the hippocampus decreases more seriously. Some studies have shown that the test repetition reproducibility of CA2-3, ca4-dg, inferior torus, and other hippocampal subregions is very similar to that of the hippocampus, which indicates that the hippocampal sub-region can also be used to reflect or predict disease progression (16). The hippocampal sub-region may be more suitable for predicting AD. The amygdala, which is attached to the end of the hippocampus, is an important brain structure for emotion, learning, and memory. In the medium brain, the entorhinal cortex connects the neocortex with the hippocampal formation. The primary excitatory source for hippocampal formation is the perforating bundle from the neurons in the second and third layers of the entorhinal cortex. Conversely, the neurons in the fourth layer receive most of the hippocampal efferent projections (17). When AD occurs, the neurons in the second and fourth layers of the entorhinal cortex are involved, resulting in the interruption of the afferent/efferent pathway of the hippocampal formation (18). The hippocampus, hippocampal subregion, entorhinal cortex, and temporal lobe are used as independent indicators to study AD progression. In the temporal lobe, there are sensory language centers located in the posterior part of the superior temporal gyrus, auditory centers located in the middle part of the superior temporal gyrus and transverse temporal gyrus, olfactory centers located in the anterior part of the hippocampal gyrus of sulcus gyrus, and advanced centers related to memory and association (19).
Conversely, the hippocampus is located on the medial side of the temporal lobe. Thus, temporal lobe atrophy can be used to predict the progress of AD. The inferior horn of the lateral ventricle has a particular anatomical structure, extending forwards and downwards into the temporal lobe (20). When AD occurs, brain parenchyma atrophy, ventricular enlargement, and ventricular volume enlargement can be used as characteristic indexes to predict AD. Thus, with the passage of the disease from MCI to the AD, the index decreases, and the degree of brain atrophy becomes more and more serious. Some studies have applied the phenomenon of asymmetry in the cerebral hemisphere of AD to drug research (21). In future disease treatments and drug research, the choice of drug delivery mode and drug concentration should be different between the left and right hemispheres of the brain to get the best clinical effects (22).
MCI is divided into EMCI and LMCI. If a correct judgment of the disease is made in the early stage of AD, the burden that the disease places on society, families, and individuals will be significantly reduced. In this study, CN-EMCI-LMCI-AD were classified in pairs to increase the specificity and detail of the classification and to enable the early diagnoses of the disease. The three classification models used in this study (i.e., RF, SVM and DT) had the highest accuracy in CN-AD, followed by EMCI-AD, CN-LMCI, LMCI-AD, and EMCI-LMCI. The accuracy of CN-EMCI prediction was low. However, in general, the accuracy of the six groups was relatively high, especially for EMCI-AD. After introducing gender and age, the accuracy rate of CN-AD was 91.07%. Based on the principal component analysis (PCA)-Fisher linear discriminant analysis (FLDA) ensemble classifier, CN-EMCI-LMCI-AD were classified in pairs, and the classification results of CN-AD and CN-EMCI were 95.65% and 60.53%, respectively (23). CN-AD had the highest prediction accuracy, which may be due to the brain differences between the NC group and AD group. EMCI and LMCI represent the middle stage of disease progression. Specifically, EMCI is the early stage of MCI, and LMCI is in the late stage of MCI. Thus, the prediction accuracy of CN-EMCI and CN-LMCI was lower than that of CN-AD. The accuracy of CN-EMCI classification was the lowest among all the indexes. The accuracy of CN-EMCI classification was very low; the highest CN-EMCI classification was 60.53%, and the overall accuracy of CN-EMCI classification in this study was 77.78% (24). Thus, the prediction accuracy of CN-EMCI should be improved to enable diagnosis at the earliest stage of the disease, implement the corresponding prevention and control measures, and prevent the occurrence and progress of the disease. If the sample size were increased, it could be used in a four-classification study of CN-EMCI-LMCI-AD.
Machine learning research uses computer training data to produce a “model”. Machine learning has been gradually maturing and has been applied to the classification and prediction of AD. The RF classifier is an algorithm model. The advantage of the RF is that it had a good performance in the data set. Due to the randomness of the RF was not easy to overfit, and it also had the advantage of anti-noise ability. Further, it can process both discrete data and continuous data and has unique advantages in the processing of high-dimensional data, including its simple and easy implementation, and low computational overhead. The RF uses the bootstrap resampling method to extract samples from the original samples, it then models each sample, and then votes on all DTs to select the final result. As RF is a classifier based on DTs, a DT classifier was also used as a classification prediction model in this study and compared to the RF.
At present, the SVM is a popular prediction model to predict AD. The highest accuracy rate of the SVM for the classification and prediction of AD is 91.6% (25), and the lowest is only 59.1%. Some research (26,27) has used the SVM for the classification and prediction of CN-AD, CN-MCI, and MCI-AD, for which the accuracy rates were 89%, 79%, and 85%, respectively. This study examined 3 classification prediction models, and the optimal model was selected following comparisons. The results showed that all MRI features, including the 272 raw data and the five indexes, SV of SA, CV, TA, and HS had the best classification and prediction effects in the RF classifier, followed by the SVM. Conversely, the DT had the worst comprehensive prediction effect. The classification accuracy and AUC of the latter were higher than the former, and similar results were found for EMCI-AD, CN-LMCI, LMCI-AD, EMCI-LMCI, CN-EMCI classifications. The best prediction model was the RF classification prediction model, which was based on five MRI indexes. Thus, RF can be used to predict MCI/AD. If the sample size was increased, we could also predict the risk of MCI transforming into AD.
In conclusion, compared to different machine learning classifiers, the RF classifier was the best algorithm to predict the progression of AD. Compared to additional MRI features, five MRI features were the best for classification. Specifically, in terms of classification accuracy, CN-AD was the best, followed by EMCI-AD, CN-LMCI, LMCI-AD, EMCI-LMCI, and CN-EMCI. Overall, the classification accuracy of the RF for CN-EMCI-LMCI-AD was higher than that of the other models. The RF classifier can effectively improve the efficiency of MCI/AD automatic diagnosis, and can classify the different stages of AD in the early stage to assist in diagnosing AD.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-2013
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-2013). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Massa F, Meli R, Morbelli S, et al. Serum neurofilament light chain rate of change in Alzheimer’s disease: potentials applications and notes of caution. Ann Transl Med 2019;7:S133. [Crossref] [PubMed]
- Pybus M, Luisi P, Dall'Olio GM, et al. Hierarchical boosting: a machine-learning framework to detect and classify hard selective sweeps in human populations. Bioinformatics 2015;31:3946-52. [Crossref] [PubMed]
- Polson NG, Scott JG, Willard BT. Proximal algorithms in statistics and machine learning. Statistical Science 2015;30:559-81. [Crossref]
- Sukumaran J, Economo EP, Lacey Knowles L. Machine learning biogeographic processes from biotic patterns: a new trait-dependent dispersal and diversification model with model choice by simulation-trained discriminant analysis. Syst Biol 2016;65:525-45. [Crossref] [PubMed]
- Zibar D, Piels M, Jones R, et al. Machine learning techniques in optical communication. J Lightwave Technol 2015;34:1442-52. [Crossref]
- Botu V, Ramprasad R. Adaptive machine learning framework to accelerate ab initio molecular dynamics. Int J Quantum Chem 2015;115:1074-83. [Crossref]
- Hansen K, Biegler F, Ramakrishnan R, et al. Machine learning predictions of molecular properties: accurate many-body potentials and nonlocality in chemical space. J Phys Chem Lett 2015;6:2326-31. [Crossref] [PubMed]
- Sperschneider J, Gardiner DM, Dodds PN, et al. EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol 2016;210:743-61. [Crossref] [PubMed]
- Ling J, Templeton J. Evaluation of machine learning algorithms for prediction of regions of high Reynolds averaged Navier Stokes uncertainty. Phys Fluids 2015;27:085103 [Crossref]
- Goetz JN, Brenning A, Petschko H, et al. Evaluating machine learning and statistical prediction techniques for landslide susceptibility modeling. Computers & Geosciences 2015;81:1-11. [Crossref]
- Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 2015;77:43-51. [Crossref] [PubMed]
- Athey S, Imbens GW. Machine learning for estimating heterogeneous causal effects. 2015. Available online: https://ideas.repec.org/p/ecl/stabus/3350.html
- Carrasquilla J, Melko RG. Machine learning phases of matter. Nat Phys 2017;13:431-4. [Crossref]
- Raissi M, Karniadakis GE. Hidden physics models: machine learning of nonlinear partial differential equations. J Comput Phys 2018;357:125-41. [Crossref]
- Deo RC. Machine Learning in medicine. Circulation 2015;132:1920-30. [Crossref] [PubMed]
- Jean N, Burke M, Xie M, et al. Combining satellite imagery and machine learning to predict poverty. Science 2016;353:790-4. [Crossref] [PubMed]
- Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet 2015;16:321-32. [Crossref] [PubMed]
- Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science 2015;349:255-60. [Crossref] [PubMed]
- Sidiropoulos ND, De Lathauwer L, Fu X, et al. Tensor decomposition for signal processing and machine learning. IEEE Trans Signal Process 2017;65:3551-82. [Crossref]
- Guzior N, Wieckowska A, Panek D, et al. Recent development of multifunctional agents as potential drug candidates for the treatment of Alzheimer's disease. Curr Med Chem 2015;22:373-404. [Crossref] [PubMed]
- Koch K, Myers NE, Göttler J, et al. Disrupted intrinsic networks link amyloid-β pathology and impaired cognition in prodromal Alzheimer's disease. Cereb Cortex 2015;25:4678-88. [Crossref] [PubMed]
- Young JE, Boulanger-Weill J, Williams DA, et al. Elucidating molecular phenotypes caused by the SORL1 Alzheimer's disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell 2015;16:373-85. [Crossref] [PubMed]
- Verdile G, Keane KN, Cruzat VF, et al. Inflammation and oxidative stress: the molecular connectivity between insulin resistance, obesity, and Alzheimer’s disease. Mediators of Inflammation 2015;2015:105828 [Crossref] [PubMed]
- Liang Y, Pertzov Y, Nicholas JM, et al. Visual short-term memory binding deficit in familial Alzheimer's disease. Cortex 2016;78:150-64. [Crossref] [PubMed]
- Fiala M, Liu QN, Sayre J, et al. Cyclooxygenase-2-positive macrophages infiltrate the Alzheimer's disease brain and damage the blood-brain barrier. Eur J Clin Invest 2002;32:360-71. [Crossref] [PubMed]
- Ossenkoppele R, Pijnenburg YA, Perry DC, et al. The behavioural/dysexecutive variant of Alzheimer's disease: clinical, neuroimaging and pathological features. Brain 2015;138:2732-49. [Crossref] [PubMed]
- Gjoneska E, Pfenning AR, Mathys H, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature 2015;518:365-9. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


